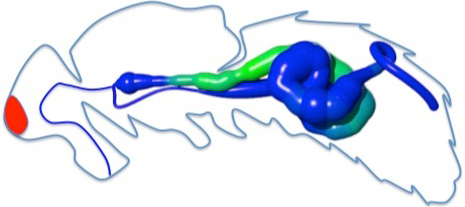
| Home | Overview of gut regions | Anatomy | Histology | Transgene expression mapping | Gene expression |
Mean expression enrichment by gut region
| Filter out everything which GO terms when the expression scale ratio is comprised between -1.5 and 1.5 for all regions |
| GO term | Number of genes | Crop / Full gut |
R2 / Full gut |
R3 / Full gut |
R4 / Full gut |
R5 / Full gut |
Hindgut / Full gut |
Cardia/R1 / Full gut |
|---|---|---|---|---|---|---|---|---|
| exocyst | 6 | -1.08 | 1.29 | 1.41 | 1.24 | 1.37 | 1.05 | 1.07 |
| M phase | 379 | 1.18 | 1.09 | 1.2 | 1.09 | 1.24 | 1.18 | 1.08 |
| protein deneddylation | 3 | 1.57 | -1.01 | 1.18 | 1.14 | 1.14 | 1.07 | -1.1 |
| cell morphogenesis | 428 | 1.11 | 1.09 | 1.24 | 1.04 | 1.25 | 1.24 | 1.24 |
| stress fiber | 1 | 1.76 | 1.07 | 1.13 | 1.08 | 1.49 | 1.28 | 1.15 |
| glycoprotein binding | 1 | 3.21 | -1.21 | 1.67 | 1.12 | 1.32 | 6.4 | 2.23 |
| lymphocyte mediated immunity | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| renal sodium ion transport | 1 | -52.82 | -1.7 | -31.49 | 1.13 | -9.63 | 16.55 | -13.85 |
| molecular_function | 8833 | 1.01 | -1.01 | 1.0 | 1.01 | 1.05 | 1.02 | -1.0 |
| ferrochelatase activity | 1 | 1.62 | 1.26 | 1.09 | -1.07 | 1.01 | 1.21 | 1.32 |
| mesoderm development | 91 | 1.26 | -1.02 | 1.08 | -1.05 | 1.17 | 1.32 | 1.03 |
| RNA splicing | 186 | 1.18 | 1.12 | 1.22 | 1.22 | 1.31 | 1.16 | 1.02 |
| nicotinamidase activity | 1 | -2.04 | 1.72 | -1.29 | 1.07 | 1.25 | 4.26 | 2.23 |
| lipoate biosynthetic process | 2 | 1.65 | 1.07 | 1.42 | 1.08 | 1.12 | 2.67 | -1.13 |
| FMN binding | 10 | 1.36 | 1.09 | 1.23 | 1.12 | -1.05 | 1.11 | -1.08 |
| response to UV-B | 1 | 1.85 | -1.14 | 1.04 | 1.61 | 1.1 | 1.04 | 1.04 |
| ectodermal cell differentiation | 2 | 4.98 | -1.19 | 1.75 | -1.46 | 1.16 | 1.31 | 1.35 |
| vesicle membrane | 23 | -1.12 | 1.13 | 1.13 | 1.07 | 1.16 | 1.18 | 1.23 |
| nuclear matrix | 3 | -1.0 | 1.28 | 1.38 | 1.16 | 1.24 | 1.52 | 1.32 |
| ceramidase activity | 2 | -7.56 | -2.15 | -1.17 | 1.28 | -1.14 | -15.17 | -6.93 |
| acyl-CoA delta11-desaturase activity | 1 | -3.53 | -3.34 | -2.71 | -4.29 | -3.06 | -2.31 | 4.71 |
| peptidyl-serine modification | 3 | 1.28 | 1.07 | 1.17 | 1.15 | 1.14 | 1.08 | -1.08 |
| cytokine binding | 3 | 1.17 | 1.12 | 1.1 | 1.02 | 1.54 | 1.2 | 1.72 |
| T-tubule | 1 | 1.33 | 1.32 | 2.26 | 1.11 | 1.52 | 1.39 | 1.48 |
| autosome | 1 | 1.07 | 1.1 | 1.33 | 1.13 | 1.37 | -1.12 | 1.05 |
| A band | 9 | 1.95 | -1.09 | -1.15 | -1.11 | 1.13 | 1.79 | 1.07 |
| cleavage furrow | 9 | -1.04 | 1.09 | 1.57 | 1.01 | 1.45 | 1.33 | 1.15 |
| macromolecule localization | 473 | 1.01 | 1.1 | 1.21 | 1.1 | 1.24 | 1.12 | 1.09 |
| ESCRT-0 complex | 2 | -1.75 | 1.13 | 2.24 | 1.06 | 1.39 | 1.34 | 1.26 |
| ribosome localization | 1 | 2.15 | -1.01 | 1.44 | 1.25 | 1.44 | 1.28 | 1.21 |
| protein localization in Golgi apparatus | 10 | -1.16 | 1.12 | 1.17 | 1.04 | 1.21 | 1.41 | 1.65 |
| pupation | 4 | 2.01 | -1.05 | 1.24 | -1.24 | 1.14 | 2.15 | 1.25 |
| DNA dealkylation | 1 | 1.12 | -1.01 | 1.19 | 1.74 | 1.2 | 1.06 | -1.09 |
| AP-2 adaptor complex binding | 1 | 1.51 | 1.33 | 2.01 | 1.25 | 1.81 | 2.91 | 1.9 |
| cargo receptor activity | 36 | 1.1 | -1.14 | -1.08 | -1.02 | -1.03 | 1.05 | -1.08 |
| regulation of growth | 153 | 1.13 | 1.15 | 1.39 | 1.07 | 1.32 | 1.3 | 1.24 |
| DNA protection | 1 | 1.02 | -1.01 | 1.31 | 1.02 | 1.58 | 1.3 | 1.2 |
| presynaptic membrane | 8 | -1.25 | 1.03 | 1.27 | -1.02 | 1.34 | 1.8 | 1.18 |
| fibril | 1 | 1.02 | -1.0 | 1.14 | 1.18 | 1.1 | 1.03 | -1.08 |
| DNA hypermethylation | 3 | 1.11 | 1.01 | 1.05 | 1.05 | 1.09 | 1.06 | -1.09 |
| main axon | 2 | -1.07 | -1.38 | -1.02 | -1.09 | 1.18 | 3.54 | 1.59 |
| actin nucleation | 4 | -2.38 | 1.35 | 1.23 | 1.09 | 1.22 | -1.94 | 1.21 |
| negative regulation of S phase of mitotic cell cycle | 11 | 1.34 | 1.21 | 1.41 | 1.15 | 1.44 | 1.65 | 1.53 |
| imaginal disc fusion | 16 | 1.01 | 1.24 | 1.46 | 1.12 | 1.3 | 1.23 | 1.22 |
| female sex differentiation | 19 | 1.28 | 1.1 | 1.09 | 1.01 | 1.21 | 1.14 | 1.12 |
| protein autophosphorylation | 7 | -1.02 | 1.18 | 1.21 | 1.11 | 1.22 | 1.16 | 1.21 |
| habituation | 4 | 1.3 | -1.01 | 1.62 | 1.18 | 1.4 | 1.83 | 1.26 |
| eye pigmentation | 34 | 1.13 | -1.1 | -1.23 | 1.11 | 1.01 | -1.18 | -1.07 |
| chromosome localization | 18 | 1.21 | 1.05 | 1.2 | 1.17 | 1.42 | 1.12 | -1.0 |
| localization | 1626 | -1.02 | 1.02 | 1.05 | 1.01 | 1.11 | 1.1 | 1.04 |
| actin crosslink formation | 1 | 2.2 | 1.13 | -1.01 | 1.18 | 1.16 | -1.08 | 1.17 |
| glycolipid binding | 1 | -2.99 | 1.35 | -1.24 | 1.41 | -1.1 | -2.55 | -1.06 |
| catecholamine transport | 2 | 3.07 | -1.44 | -1.47 | -1.28 | -1.45 | -1.2 | -1.42 |
| carboxylic ester hydrolase activity | 99 | -1.69 | -1.12 | -1.47 | -1.07 | -1.31 | -2.03 | -1.33 |
| foraging behavior | 1 | -12.04 | -1.69 | 9.34 | -2.59 | -11.09 | -12.8 | -6.11 |
| regulation of canonical Wnt receptor signaling pathway | 6 | 1.19 | 1.08 | 1.68 | 1.07 | 1.07 | 1.06 | 1.46 |
| establishment of blood-brain barrier | 19 | 2.5 | -1.26 | 1.24 | -1.17 | 1.21 | 2.3 | 1.4 |
| neuroendocrine cell differentiation | 2 | -1.09 | 1.14 | 1.11 | 1.1 | 1.07 | -1.24 | 1.07 |
| Wnt protein secretion | 1 | 1.86 | -1.05 | 2.03 | 1.04 | 1.26 | 2.03 | 1.19 |
| vitamin B6 binding | 40 | 1.01 | 1.02 | -1.27 | -1.04 | -1.03 | -1.02 | 1.04 |
| RISC-loading complex | 1 | 1.44 | 1.23 | 1.89 | 1.19 | 2.25 | 1.64 | 1.56 |
| cellular response to ecdysone | 11 | 1.32 | 1.19 | 1.43 | 1.13 | 1.47 | 1.53 | 1.33 |
| circulatory system development | 70 | 1.28 | -1.02 | 1.19 | -1.07 | 1.17 | 1.3 | 1.17 |
| regulation of ion homeostasis | 3 | 1.14 | 1.14 | 1.39 | 1.16 | 1.04 | 1.71 | 1.47 |
| mitochondrial genome maintenance | 8 | -1.05 | 1.17 | 1.44 | 1.18 | 1.23 | 1.21 | -1.27 |
| reproduction | 846 | 1.12 | 1.06 | 1.15 | 1.03 | 1.18 | 1.14 | 1.14 |
| ribosome biogenesis | 52 | 1.28 | 1.16 | 1.24 | 1.3 | 1.29 | 1.06 | 1.05 |
| protein binding involved in protein folding | 2 | 1.49 | 1.24 | 1.02 | 1.11 | 1.08 | 1.24 | 1.18 |
| unfolded protein binding | 80 | 1.1 | 1.04 | 1.1 | 1.14 | 1.14 | 1.02 | -1.05 |
| protein thiol-disulfide exchange | 1 | -1.26 | 1.49 | 1.56 | 1.08 | 1.23 | -1.31 | 1.33 |
| disulfide oxidoreductase activity | 30 | 1.14 | 1.05 | -1.02 | 1.16 | 1.09 | -1.11 | -1.1 |
| alpha-1,6-mannosyltransferase activity | 1 | 1.2 | 1.3 | 1.28 | 1.05 | 1.27 | 1.14 | 1.6 |
| trans-hexaprenyltranstransferase activity | 2 | 2.38 | 1.18 | 1.12 | 1.03 | 1.43 | 1.86 | 1.15 |
| single strand break repair | 2 | 1.23 | 1.1 | 1.14 | 1.12 | 1.12 | 1.05 | 1.04 |
| single-stranded DNA specific endodeoxyribonuclease activity | 3 | 1.42 | 1.05 | 1.38 | 1.12 | 1.4 | 1.2 | -1.23 |
| phosphopyruvate hydratase complex | 1 | 1.97 | 1.06 | -1.39 | -1.17 | -1.32 | 2.59 | 1.17 |
| alpha-glucoside transport | 2 | 1.24 | -2.16 | -1.19 | -2.0 | -1.13 | 3.38 | 1.3 |
| regulation of DNA recombination | 2 | -1.59 | 1.15 | 1.13 | 1.19 | 1.42 | -1.74 | -1.31 |
| regulation of mitotic recombination | 1 | -2.02 | 1.1 | 1.06 | 1.2 | 1.9 | -2.66 | -1.77 |
| negative regulation of mitotic recombination | 1 | -2.02 | 1.1 | 1.06 | 1.2 | 1.9 | -2.66 | -1.77 |
| mitotic spindle elongation | 72 | 1.4 | 1.08 | 1.2 | 1.04 | 1.24 | 1.24 | 1.15 |
| alpha-1,2-mannosyltransferase activity | 1 | -1.58 | 1.15 | -1.08 | 1.11 | -1.2 | -1.77 | 1.49 |
| mannosyltransferase activity | 12 | -1.24 | 1.17 | 1.3 | 1.07 | 1.33 | -1.21 | 1.12 |
| acyl carrier activity | 6 | 1.59 | 1.03 | 1.12 | 1.01 | 1.14 | 1.4 | 1.17 |
| very long-chain fatty acid metabolic process | 5 | 3.63 | -2.39 | -2.2 | -3.1 | -1.91 | -1.08 | 3.71 |
| long-chain fatty acid transporter activity | 7 | -2.15 | 1.3 | -1.32 | -1.52 | -2.35 | -1.39 | -1.5 |
| plasma membrane | 575 | 1.16 | 1.01 | 1.11 | -1.03 | 1.11 | 1.23 | 1.16 |
| transition metal ion transport | 17 | 1.11 | 1.0 | 1.11 | -1.05 | -1.09 | 1.21 | 1.05 |
| protein targeting to Golgi | 10 | -1.16 | 1.12 | 1.17 | 1.04 | 1.21 | 1.41 | 1.65 |
| autophagic vacuole assembly | 2 | 1.21 | 1.14 | -1.0 | 1.38 | 1.03 | -1.03 | -1.25 |
| autophagic vacuole fusion | 2 | 1.14 | 1.16 | 1.85 | 1.13 | 1.26 | 1.5 | 1.09 |
| electron carrier activity | 154 | -1.07 | -1.09 | -1.18 | -1.02 | 1.01 | -1.08 | -1.1 |
| tRNA binding | 9 | 1.2 | 1.27 | 1.24 | 1.22 | 1.15 | 1.15 | 1.01 |
| urea cycle | 1 | -31.89 | 1.77 | -20.72 | 1.41 | 1.0 | -12.66 | -3.5 |
| argininosuccinate metabolic process | 1 | -31.89 | 1.77 | -20.72 | 1.41 | 1.0 | -12.66 | -3.5 |
| ribosomal subunit export from nucleus | 1 | 2.15 | -1.01 | 1.44 | 1.25 | 1.44 | 1.28 | 1.21 |
| ribosomal large subunit export from nucleus | 1 | 2.15 | -1.01 | 1.44 | 1.25 | 1.44 | 1.28 | 1.21 |
| protein import into nucleus, docking | 2 | -1.85 | 1.21 | 1.23 | 1.37 | 1.53 | -1.37 | -1.23 |
| protein import into nucleus, translocation | 2 | 1.98 | -1.11 | 1.05 | 1.22 | 1.33 | 2.03 | -1.05 |
| fatty-acyl-CoA binding | 8 | -2.25 | -1.27 | -1.85 | 1.18 | 1.35 | -2.52 | -1.44 |
| L-ornithine transmembrane transporter activity | 1 | 1.32 | 1.45 | -1.5 | 1.71 | 1.05 | 1.03 | -1.1 |
| mitochondrial ornithine transport | 1 | 1.32 | 1.45 | -1.5 | 1.71 | 1.05 | 1.03 | -1.1 |
| DNA replication | 73 | 1.08 | 1.13 | 1.19 | 1.17 | 1.34 | 1.13 | -1.02 |
| chromosome segregation | 124 | 1.15 | 1.1 | 1.25 | 1.11 | 1.3 | 1.2 | 1.1 |
| chromosome organization | 265 | 1.2 | 1.11 | 1.27 | 1.13 | 1.32 | 1.2 | 1.15 |
| mitotic sister chromatid segregation | 55 | 1.25 | 1.07 | 1.29 | 1.1 | 1.36 | 1.27 | 1.09 |
| M phase specific microtubule process | 1 | 1.59 | 1.65 | -1.13 | 1.13 | 1.0 | -1.46 | -1.04 |
| cell cycle checkpoint | 84 | 1.23 | 1.14 | 1.35 | 1.19 | 1.41 | 1.29 | 1.15 |
| DNA replication checkpoint | 3 | -1.03 | -1.02 | 1.14 | 1.12 | 1.37 | 1.02 | -1.1 |
| DNA damage checkpoint | 69 | 1.23 | 1.15 | 1.38 | 1.19 | 1.41 | 1.31 | 1.15 |
| regulation of cyclin-dependent protein kinase activity | 15 | 1.32 | 1.16 | 1.36 | 1.2 | 1.41 | 1.07 | 1.15 |
| G1 phase of mitotic cell cycle | 1 | -1.2 | 1.72 | 1.55 | 1.31 | 1.53 | 1.21 | 1.72 |
| G1/S transition of mitotic cell cycle | 14 | 1.35 | 1.24 | 1.43 | 1.21 | 1.54 | 1.24 | 1.25 |
| regulation of transcription involved in G1/S phase of mitotic cell cycle | 1 | 1.31 | 1.43 | 1.45 | 1.16 | 1.14 | 1.54 | 1.46 |
| S phase of mitotic cell cycle | 3 | -1.1 | 1.13 | 1.05 | 1.24 | 1.2 | -1.11 | -1.1 |
| astral microtubule | 4 | -1.02 | 1.17 | 1.36 | -1.01 | 1.32 | 1.27 | 1.24 |
| G2 phase of mitotic cell cycle | 1 | 1.67 | 1.26 | 1.32 | 1.4 | 1.23 | 1.13 | 1.21 |
| G2/M transition of mitotic cell cycle | 71 | 1.21 | 1.14 | 1.34 | 1.19 | 1.41 | 1.24 | 1.13 |
| M phase of mitotic cell cycle | 148 | 1.21 | 1.1 | 1.27 | 1.12 | 1.33 | 1.23 | 1.1 |
| mitotic metaphase | 1 | -1.06 | 1.01 | -1.03 | 1.11 | -1.07 | -1.08 | -1.08 |
| mitotic anaphase | 19 | 1.3 | 1.13 | 1.29 | 1.17 | 1.36 | 1.32 | 1.11 |
| mitotic anaphase A | 4 | -1.13 | 1.29 | 1.25 | 1.15 | 1.37 | 1.13 | 1.24 |
| mitotic anaphase B | 2 | 1.04 | 1.25 | 1.4 | 1.16 | 1.52 | -1.16 | 1.12 |
| mitotic telophase | 1 | 1.4 | 1.02 | 1.15 | 1.14 | 1.72 | -1.18 | -1.16 |
| sulfur amino acid metabolic process | 11 | 1.06 | -1.1 | -1.32 | 1.02 | -1.36 | 1.08 | -1.14 |
| sulfur amino acid biosynthetic process | 3 | 1.32 | 1.11 | 1.04 | -1.09 | -1.92 | 1.19 | 1.3 |
| sulfur amino acid catabolic process | 1 | 1.48 | 1.28 | 1.32 | 1.05 | 1.07 | 1.16 | -1.01 |
| sulfur amino acid transmembrane transporter activity | 1 | -7.18 | -1.45 | -9.47 | 1.62 | 3.49 | -2.41 | -5.84 |
| sulfur amino acid transport | 1 | -7.18 | -1.45 | -9.47 | 1.62 | 3.49 | -2.41 | -5.84 |
| sulfate assimilation | 1 | -2.63 | 1.06 | 1.41 | 1.07 | 1.11 | -1.64 | -1.02 |
| succinate dehydrogenase activity | 6 | 1.02 | 1.03 | 1.28 | -1.03 | -1.0 | 1.38 | -1.21 |
| nucleotide-excision repair complex | 3 | 1.73 | 1.2 | 1.21 | 1.15 | 1.32 | 1.1 | -1.16 |
| regulation of transcription involved in G2/M-phase of mitotic cell cycle | 1 | 1.12 | 1.4 | 1.69 | -1.0 | 1.67 | 1.7 | 1.71 |
| histone deacetylase complex | 16 | 1.3 | 1.16 | 1.3 | 1.17 | 1.34 | 1.37 | 1.21 |
| negative regulation of transcription from RNA polymerase II promoter | 104 | 1.12 | 1.05 | 1.08 | -1.01 | 1.16 | 1.18 | 1.11 |
| histone acetyltransferase complex | 40 | 1.37 | 1.14 | 1.22 | 1.23 | 1.26 | 1.13 | 1.06 |
| SAGA complex | 20 | 1.38 | 1.1 | 1.14 | 1.26 | 1.22 | 1.03 | -1.02 |
| transcription factor TFIIIB complex | 2 | 1.19 | 1.29 | 1.5 | 1.13 | 1.51 | 1.31 | 1.18 |
| establishment of mitotic spindle orientation | 2 | 1.02 | 1.4 | 1.82 | 1.21 | 1.49 | 1.4 | 1.34 |
| Golgi cis cisterna | 1 | -1.12 | 1.19 | 1.36 | 1.12 | 1.23 | -1.28 | 1.74 |
| Golgi trans cisterna | 2 | -1.03 | 1.16 | 1.1 | 1.38 | 1.81 | 1.12 | 1.1 |
| Golgi membrane | 37 | 1.04 | 1.11 | 1.07 | 1.1 | 1.06 | 1.29 | 1.38 |
| microfilament motor activity | 3 | 6.28 | -1.75 | 1.12 | -1.66 | 1.09 | 3.74 | -1.21 |
| SNARE binding | 6 | -1.14 | 1.05 | 1.42 | 1.11 | 1.29 | 1.2 | 1.24 |
| recombinase activity | 4 | 1.15 | 1.04 | -1.05 | 1.11 | 1.09 | -1.16 | -1.08 |
| ubiquitin ligase complex | 43 | 1.31 | 1.15 | 1.31 | 1.16 | 1.28 | 1.18 | 1.18 |
| nuclear ubiquitin ligase complex | 19 | 1.18 | 1.15 | 1.35 | 1.18 | 1.34 | 1.11 | 1.14 |
| cytoplasmic ubiquitin ligase complex | 1 | 2.69 | 1.18 | 1.72 | 1.15 | -1.01 | -1.18 | -1.08 |
| rRNA modification | 5 | 1.57 | 1.18 | 1.26 | 1.33 | 1.41 | 1.18 | -1.02 |
| two-component sensor activity | 3 | 1.12 | -1.13 | 1.03 | -1.08 | 1.69 | 1.52 | -1.14 |
| two-component response regulator activity | 1 | -1.1 | 1.39 | 1.36 | 1.05 | 1.38 | 3.53 | 1.68 |
| protein phosphatase type 2A complex | 8 | 1.09 | 1.04 | 1.37 | 1.07 | 1.23 | 2.15 | 1.1 |
| two-component signal transduction system (phosphorelay) | 3 | -1.1 | 1.14 | 1.08 | -1.02 | -1.02 | 1.53 | 1.01 |
| protein phosphatase type 1 complex | 5 | 1.43 | 1.05 | 1.28 | 1.01 | 1.16 | 1.5 | 1.4 |
| MAPK cascade | 35 | -1.01 | 1.14 | 1.46 | 1.09 | 1.46 | 1.19 | 1.31 |
| nucleotide binding | 1153 | 1.12 | 1.07 | 1.11 | 1.06 | 1.16 | 1.16 | 1.09 |
| ribonuclease MRP complex | 1 | 1.84 | 1.03 | 1.1 | 1.42 | 1.17 | 1.05 | -1.14 |
| 3'-5'-exoribonuclease activity | 15 | 1.46 | 1.13 | 1.16 | 1.27 | 1.25 | 1.2 | 1.04 |
| nuclear exosome (RNase complex) | 13 | 1.3 | 1.17 | 1.14 | 1.29 | 1.27 | 1.13 | 1.05 |
| cytoplasmic exosome (RNase complex) | 8 | 1.45 | 1.11 | 1.16 | 1.3 | 1.29 | 1.15 | -1.0 |
| exosome (RNase complex) | 13 | 1.3 | 1.17 | 1.14 | 1.29 | 1.27 | 1.13 | 1.05 |
| rRNA (adenine-N6,N6-)-dimethyltransferase activity | 2 | 1.61 | 1.27 | 1.44 | 1.22 | 1.57 | 1.46 | 1.13 |
| cytosolic large ribosomal subunit | 50 | 1.38 | 1.02 | 1.09 | 1.0 | 1.14 | 1.2 | 1.09 |
| cytosolic small ribosomal subunit | 33 | 1.37 | 1.02 | 1.1 | 1.02 | 1.14 | 1.17 | 1.04 |
| rDNA binding | 2 | -1.04 | 1.15 | 1.82 | 1.29 | 2.02 | 1.26 | 1.15 |
| nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 12 | -1.19 | 1.17 | 1.13 | 1.22 | 1.25 | -1.02 | 1.0 |
| activation of MAPKKK activity | 1 | -1.25 | 1.21 | 1.4 | 1.19 | 1.07 | 1.11 | 1.44 |
| activation of MAPKK activity | 3 | 1.19 | 1.2 | 1.4 | 1.2 | 1.34 | 1.15 | 1.19 |
| activation of MAPK activity | 7 | 1.11 | 1.21 | 1.54 | 1.12 | 1.37 | 2.0 | 1.14 |
| inactivation of MAPK activity | 1 | 2.84 | 1.44 | 1.69 | 1.19 | 1.68 | 4.37 | 1.83 |
| MAPK import into nucleus | 1 | 1.14 | 1.19 | 1.6 | 1.04 | 1.48 | 1.45 | 1.57 |
| protein binding | 1244 | 1.17 | 1.05 | 1.21 | 1.04 | 1.22 | 1.2 | 1.14 |
| modification-dependent protein catabolic process | 94 | 1.17 | 1.13 | 1.33 | 1.13 | 1.3 | 1.1 | 1.12 |
| protein ubiquitination involved in ubiquitin-dependent protein catabolic process | 5 | 1.02 | 1.13 | 2.15 | 1.07 | 1.56 | 1.16 | 1.11 |
| meiotic spindle organization | 22 | 1.17 | -1.06 | -1.01 | -1.07 | 1.19 | 1.02 | 1.17 |
| tRNA-intron endonuclease activity | 1 | 1.37 | 1.2 | -1.07 | 1.24 | 1.11 | -1.16 | -1.02 |
| tRNA-intron endonuclease complex | 1 | 1.37 | 1.2 | -1.07 | 1.24 | 1.11 | -1.16 | -1.02 |
| DNA secondary structure binding | 3 | -1.08 | 1.33 | 1.37 | 1.18 | 1.48 | -1.09 | 1.25 |
| proton-transporting two-sector ATPase complex | 48 | -1.18 | 1.0 | 1.27 | -1.11 | -1.06 | 1.39 | -1.15 |
| vacuolar proton-transporting V-type ATPase, V0 domain | 13 | -2.32 | -1.03 | 1.35 | -1.29 | -1.38 | 1.02 | -1.29 |
| vacuolar proton-transporting V-type ATPase, V1 domain | 12 | -1.17 | 1.02 | 1.44 | -1.12 | -1.04 | 1.8 | -1.01 |
| peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity | 1 | -1.09 | 1.27 | 1.39 | 1.28 | 1.54 | -1.1 | 1.18 |
| microtubule cytoskeleton organization | 338 | 1.13 | 1.1 | 1.24 | 1.09 | 1.26 | 1.18 | 1.09 |
| nuclear chromosome | 88 | 1.19 | 1.12 | 1.23 | 1.14 | 1.27 | 1.13 | 1.03 |
| cytoplasmic chromosome | 1 | 1.18 | 1.16 | 1.75 | 1.38 | 1.68 | 1.06 | -1.24 |
| condensed nuclear chromosome | 15 | 1.25 | 1.06 | 1.23 | 1.06 | 1.25 | 1.24 | 1.09 |
| condensed chromosome | 38 | 1.15 | 1.06 | 1.23 | 1.09 | 1.3 | 1.2 | 1.07 |
| mitotic prometaphase | 1 | -1.39 | -1.16 | -1.07 | 1.34 | 1.62 | -1.16 | -1.17 |
| pericentriolar material | 6 | 1.07 | 1.05 | 1.08 | 1.04 | 1.14 | 1.08 | 1.03 |
| spliceosomal complex assembly | 14 | 1.26 | 1.11 | 1.27 | 1.23 | 1.28 | 1.21 | -1.02 |
| C-4 methylsterol oxidase activity | 2 | -1.31 | -6.89 | -7.44 | -1.07 | -3.2 | -1.65 | -1.68 |
| allantoin metabolic process | 2 | 3.32 | -1.45 | -1.35 | -1.39 | -1.17 | 1.29 | -1.34 |
| allantoin catabolic process | 1 | 2.85 | -1.01 | 1.16 | 1.03 | -1.08 | 1.74 | -1.1 |
| nitrilase activity | 2 | 1.33 | 1.42 | -1.12 | 1.25 | 1.47 | 1.28 | -1.05 |
| sodium:amino acid symporter activity | 5 | 1.26 | 1.14 | -2.06 | -1.96 | -1.76 | -1.26 | 1.36 |
| nucleoside transmembrane transporter activity | 6 | -1.51 | 1.3 | -1.37 | 1.14 | -1.09 | -1.36 | -1.05 |
| intracellular | 4036 | 1.14 | 1.07 | 1.15 | 1.08 | 1.18 | 1.16 | 1.05 |
| proton-transporting ATPase activity, rotational mechanism | 21 | 1.15 | -1.01 | 1.22 | -1.1 | -1.03 | 1.64 | -1.08 |
| mitochondrial chromosome | 1 | 1.18 | 1.16 | 1.75 | 1.38 | 1.68 | 1.06 | -1.24 |
| GTPase activity | 136 | 1.2 | 1.08 | 1.29 | 1.07 | 1.22 | 1.09 | 1.05 |
| heterotrimeric G-protein complex | 18 | 1.39 | 1.11 | 1.42 | 1.06 | 1.22 | 1.03 | 1.1 |
| mitochondrial fission | 2 | 1.47 | -1.03 | 1.65 | 1.15 | 1.07 | 1.58 | -1.11 |
| cell fraction | 106 | -1.02 | -1.12 | -1.21 | -1.02 | 1.06 | -1.09 | -1.04 |
| peroxisome targeting sequence binding | 3 | 1.2 | -1.04 | -1.07 | 1.13 | 1.12 | -1.21 | -1.06 |
| peptidoglycan metabolic process | 12 | -1.13 | -1.64 | -1.19 | -1.04 | -1.82 | -1.69 | -1.29 |
| polysaccharide biosynthetic process | 22 | 1.55 | 1.02 | -1.01 | 1.03 | 1.12 | 1.58 | 1.25 |
| polysaccharide catabolic process | 30 | -1.17 | -1.91 | -2.09 | -1.67 | -1.68 | -1.04 | 1.4 |
| mitochondrial proton-transporting ATP synthase complex, catalytic core F(1) | 8 | 1.58 | -1.08 | 1.12 | -1.1 | 1.01 | 1.92 | -1.31 |
| mitochondrial proton-transporting ATP synthase complex, coupling factor F(o) | 9 | 1.19 | 1.04 | 1.31 | 1.01 | 1.22 | 1.62 | -1.14 |
| mitotic cell cycle | 399 | 1.21 | 1.11 | 1.24 | 1.12 | 1.29 | 1.21 | 1.1 |
| nuclear division | 149 | 1.23 | 1.09 | 1.27 | 1.11 | 1.32 | 1.26 | 1.12 |
| cytokinesis after mitosis | 5 | -1.12 | 1.17 | 1.26 | 1.17 | 1.24 | -1.37 | 1.01 |
| 1-phosphatidylinositol-3-phosphate 5-kinase activity | 1 | -1.33 | 1.29 | 1.91 | 1.33 | 2.36 | 1.28 | 1.16 |
| magnesium ion binding | 46 | 1.08 | 1.06 | 1.18 | 1.05 | 1.15 | 1.38 | 1.01 |
| nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 10 | 1.48 | 1.17 | 1.74 | 1.19 | 1.61 | 1.9 | 1.56 |
| nuclear-transcribed mRNA poly(A) tail shortening | 7 | 1.62 | 1.13 | 1.74 | 1.15 | 1.59 | 2.05 | 1.42 |
| deadenylation-dependent decapping of nuclear-transcribed mRNA | 2 | 1.27 | 1.29 | 1.78 | 1.53 | 1.51 | 1.59 | 1.46 |
| nuclear-transcribed mRNA catabolic process, exonucleolytic | 1 | -1.45 | 1.76 | 1.14 | 1.42 | 1.29 | -1.3 | 1.32 |
| integral to membrane of membrane fraction | 2 | 1.74 | 1.04 | 1.6 | 1.07 | 1.43 | 2.77 | 1.46 |
| peripheral to membrane of membrane fraction | 2 | -1.06 | 1.15 | 1.33 | 1.33 | 1.33 | 1.07 | -1.07 |
| retrograde transport, vesicle recycling within Golgi | 10 | -1.16 | 1.12 | 1.17 | 1.04 | 1.21 | 1.41 | 1.65 |
| response to reactive oxygen species | 16 | 1.05 | 1.06 | 1.08 | 1.15 | 1.2 | -1.01 | -1.15 |
| response to superoxide | 2 | 1.69 | 1.17 | 1.01 | 1.09 | 1.11 | 1.41 | -1.08 |
| response to oxygen radical | 2 | 1.69 | 1.17 | 1.01 | 1.09 | 1.11 | 1.41 | -1.08 |
| cyclin-dependent protein kinase holoenzyme complex | 7 | 1.06 | 1.09 | 1.22 | 1.21 | 1.41 | -1.23 | -1.12 |
| nicotinamide-nucleotide adenylyltransferase activity | 1 | -2.52 | 1.99 | 1.32 | 1.32 | 1.09 | -1.34 | 1.43 |
| organellar ribosome | 72 | 1.04 | 1.07 | 1.23 | 1.29 | 1.25 | 1.45 | -1.55 |
| organellar small ribosomal subunit | 30 | 1.04 | 1.09 | 1.24 | 1.27 | 1.23 | 1.47 | -1.48 |
| organellar large ribosomal subunit | 44 | 1.03 | 1.05 | 1.24 | 1.3 | 1.28 | 1.46 | -1.6 |
| protein-methionine-R-oxide reductase activity | 1 | 2.04 | 1.02 | -1.15 | -1.19 | -1.3 | 1.08 | -1.25 |
| lytic vacuole | 28 | -2.58 | -1.1 | -2.35 | 1.15 | -1.08 | -2.06 | -3.03 |
| negative regulation of transposition, DNA-mediated | 1 | -1.25 | 1.2 | 1.21 | 1.19 | 1.06 | -1.14 | 1.03 |
| regulation of transposition, DNA-mediated | 1 | -1.25 | 1.2 | 1.21 | 1.19 | 1.06 | -1.14 | 1.03 |
| RNA cap binding | 13 | 1.12 | 1.12 | 1.2 | 1.04 | 1.14 | 1.12 | 1.32 |
| RNA 7-methylguanosine cap binding | 9 | 1.06 | 1.12 | 1.24 | -1.0 | 1.11 | 1.11 | 1.44 |
| THO complex | 5 | 1.18 | 1.1 | 1.39 | 1.17 | 1.27 | 1.37 | 1.09 |
| RNA splicing, via transesterification reactions | 180 | 1.17 | 1.11 | 1.21 | 1.23 | 1.31 | 1.16 | 1.0 |
| RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 177 | 1.17 | 1.11 | 1.21 | 1.23 | 1.31 | 1.15 | 1.0 |
| alternative nuclear mRNA splicing, via spliceosome | 1 | 1.52 | 1.27 | 1.61 | 1.3 | 1.51 | 2.77 | 1.58 |
| regulation of alternative nuclear mRNA splicing, via spliceosome | 52 | 1.22 | 1.17 | 1.28 | 1.16 | 1.24 | 1.42 | 1.09 |
| spliceosomal snRNP assembly | 1 | 1.14 | -1.01 | 1.12 | 1.41 | 1.03 | -1.2 | -1.43 |
| spliceosomal complex disassembly | 1 | 1.69 | 1.29 | 1.56 | 1.32 | 1.6 | 1.63 | 1.44 |
| RNA splicing, via endonucleolytic cleavage and ligation | 3 | 1.93 | 1.15 | 1.42 | 1.2 | 1.36 | 1.44 | 1.34 |
| nuclear mRNA splicing, via spliceosome | 177 | 1.17 | 1.11 | 1.21 | 1.23 | 1.31 | 1.15 | 1.0 |
| four-way junction DNA binding | 1 | -1.12 | 1.35 | 2.36 | 1.33 | 2.9 | 1.28 | 1.43 |
| Y-form DNA binding | 1 | 1.11 | 1.31 | -1.06 | 1.14 | -1.02 | -1.04 | 1.09 |
| mitochondrion degradation | 1 | 2.65 | 1.08 | 1.15 | 1.38 | 1.25 | 2.74 | 1.17 |
| DNA-directed RNA polymerase complex | 23 | 1.32 | 1.14 | 1.23 | 1.25 | 1.26 | 1.12 | 1.07 |
| core TFIIH complex | 4 | 1.37 | 1.09 | 1.37 | 1.35 | 1.33 | 1.15 | 1.06 |
| SSL2-core TFIIH complex | 4 | 1.37 | 1.09 | 1.37 | 1.35 | 1.33 | 1.15 | 1.06 |
| maturation of 2S rRNA | 1 | 1.7 | -1.05 | -1.07 | -1.09 | -1.18 | 1.33 | 1.14 |
| base pairing | 2 | 1.38 | 1.09 | 1.14 | 1.35 | 1.29 | -1.02 | -1.04 |
| base pairing with RNA | 2 | 1.38 | 1.09 | 1.14 | 1.35 | 1.29 | -1.02 | -1.04 |
| proteasome complex | 55 | 1.37 | 1.04 | 1.2 | 1.15 | 1.17 | -1.02 | 1.02 |
| embryonic axis specification | 118 | 1.23 | 1.09 | 1.29 | 1.05 | 1.24 | 1.31 | 1.25 |
| mismatch base pair DNA N-glycosylase activity | 1 | -1.08 | 1.06 | -1.04 | 1.0 | -1.17 | 1.06 | -1.14 |
| oxidized base lesion DNA N-glycosylase activity | 3 | 2.16 | -1.01 | 1.16 | 1.13 | 1.13 | 1.51 | -1.03 |
| oxidized pyrimidine base lesion DNA N-glycosylase activity | 1 | 3.75 | 1.0 | 1.19 | 1.14 | 1.11 | 1.95 | 1.06 |
| achiasmate meiosis I | 1 | -1.17 | 1.02 | -1.13 | 1.11 | -1.02 | -1.2 | -1.13 |
| meiotic DNA recombinase assembly | 1 | -1.21 | 1.44 | 1.1 | 1.2 | 1.03 | -1.18 | 1.17 |
| meiotic mismatch repair | 1 | 1.34 | 1.15 | 1.36 | 1.37 | 1.61 | 1.16 | 1.18 |
| resolution of meiotic recombination intermediates | 4 | 1.0 | 1.09 | 1.25 | 1.17 | 1.45 | 1.14 | 1.06 |
| nucleotide-excision repair, DNA damage recognition | 1 | 1.35 | 1.22 | 1.6 | 1.15 | 1.67 | 2.6 | 1.56 |
| telomere maintenance | 14 | 1.19 | 1.12 | 1.39 | 1.17 | 1.35 | 1.2 | 1.11 |
| double-strand break repair via homologous recombination | 7 | 1.12 | 1.16 | 1.16 | 1.17 | 1.22 | -1.03 | 1.11 |
| recombinational repair | 7 | 1.12 | 1.16 | 1.16 | 1.17 | 1.22 | -1.03 | 1.11 |
| non-recombinational repair | 6 | -1.06 | 1.22 | 1.37 | 1.12 | 1.33 | -1.11 | 1.14 |
| DNA recombinase assembly | 1 | -1.21 | 1.44 | 1.1 | 1.2 | 1.03 | -1.18 | 1.17 |
| DNA synthesis involved in DNA repair | 1 | 1.11 | 1.31 | -1.06 | 1.14 | -1.02 | -1.04 | 1.09 |
| strand displacement | 1 | 1.11 | 1.31 | -1.06 | 1.14 | -1.02 | -1.04 | 1.09 |
| DNA catabolic process, endonucleolytic | 4 | 1.64 | 1.12 | 1.39 | 1.15 | 1.35 | 1.4 | 1.54 |
| DNA catabolic process, exonucleolytic | 2 | -1.31 | 1.21 | 1.11 | 1.22 | 1.62 | -1.59 | -1.29 |
| DNA strand annealing activity | 2 | 1.04 | 1.17 | 1.07 | 1.09 | 1.08 | 1.05 | 1.03 |
| karyogamy | 6 | 1.05 | 1.11 | 1.23 | 1.09 | 1.44 | 1.02 | 1.13 |
| syncytium formation by plasma membrane fusion | 27 | 1.01 | 1.06 | 1.27 | -1.02 | 1.21 | 1.27 | 1.14 |
| adenyl-nucleotide exchange factor activity | 1 | -1.14 | 1.21 | -1.05 | 1.17 | 1.0 | -1.29 | -1.36 |
| chromosome, centromeric region | 54 | 1.2 | 1.07 | 1.19 | 1.11 | 1.31 | 1.15 | 1.07 |
| kinetochore | 31 | 1.18 | 1.08 | 1.16 | 1.14 | 1.33 | 1.13 | -1.0 |
| condensed chromosome kinetochore | 9 | 1.03 | 1.06 | 1.14 | 1.12 | 1.28 | -1.06 | -1.03 |
| condensed nuclear chromosome kinetochore | 1 | 1.0 | 1.07 | -1.07 | 1.19 | -1.13 | -1.28 | 1.01 |
| condensed chromosome, centromeric region | 13 | 1.09 | 1.06 | 1.24 | 1.12 | 1.37 | 1.11 | 1.02 |
| condensed nuclear chromosome, centromeric region | 7 | 1.24 | 1.07 | 1.34 | 1.12 | 1.41 | 1.38 | 1.12 |
| chromosome, telomeric region | 10 | 1.35 | 1.09 | 1.3 | 1.17 | 1.42 | 1.18 | 1.05 |
| nuclear chromosome, telomeric region | 2 | 1.89 | 1.01 | 1.39 | 1.11 | 1.79 | 1.05 | 1.11 |
| chromatin | 122 | 1.11 | 1.12 | 1.24 | 1.14 | 1.27 | 1.15 | 1.12 |
| nucleosome | 15 | -1.1 | 1.01 | 1.25 | 1.15 | 1.09 | -1.35 | -1.01 |
| nuclear nucleosome | 1 | 1.0 | 1.07 | -1.07 | 1.19 | -1.13 | -1.28 | 1.01 |
| nuclear chromatin | 49 | 1.15 | 1.15 | 1.25 | 1.15 | 1.27 | 1.14 | 1.09 |
| euchromatin | 12 | 1.14 | 1.22 | 1.34 | 1.18 | 1.46 | 1.33 | 1.26 |
| heterochromatin | 22 | 1.14 | 1.16 | 1.29 | 1.08 | 1.32 | 1.36 | 1.33 |
| synaptonemal complex | 4 | 1.04 | 1.05 | 1.03 | 1.0 | -1.02 | -1.06 | 1.06 |
| condensin complex | 8 | 1.13 | 1.05 | 1.18 | 1.06 | 1.26 | 1.27 | 1.12 |
| sex chromosome | 7 | 1.3 | 1.17 | 1.32 | 1.09 | 1.18 | 1.39 | 1.28 |
| X chromosome | 6 | 1.37 | 1.23 | 1.41 | 1.11 | 1.23 | 1.51 | 1.37 |
| Y chromosome | 1 | -1.03 | -1.14 | -1.12 | -1.01 | -1.12 | -1.2 | -1.19 |
| origin recognition complex | 7 | 1.06 | 1.15 | 1.09 | 1.19 | 1.24 | 1.01 | -1.08 |
| GINS complex | 2 | 1.77 | 1.11 | 1.3 | 1.14 | 1.18 | 1.14 | -1.06 |
| sister chromatid segregation | 56 | 1.25 | 1.07 | 1.3 | 1.11 | 1.37 | 1.27 | 1.09 |
| inositol-1,4,5-trisphosphate 6-kinase activity | 1 | -1.15 | 1.56 | -1.07 | 1.07 | -1.15 | -1.25 | 1.09 |
| inositol hexakisphosphate kinase activity | 1 | -1.38 | 1.16 | 2.02 | -1.07 | 2.12 | 1.33 | 1.59 |
| translation repressor activity, nucleic acid binding | 6 | 1.94 | 1.04 | 2.03 | 1.04 | 1.61 | 1.46 | 1.82 |
| cell morphogenesis involved in differentiation | 351 | 1.14 | 1.09 | 1.26 | 1.04 | 1.28 | 1.28 | 1.26 |
| cytokinesis | 81 | 1.03 | 1.07 | 1.23 | 1.06 | 1.32 | 1.09 | 1.11 |
| assembly of actomyosin apparatus involved in cell cycle cytokinesis | 14 | 1.26 | -1.24 | -1.03 | -1.27 | 1.08 | 1.13 | 1.08 |
| cytokinesis, actomyosin contractile ring assembly | 14 | 1.26 | -1.24 | -1.03 | -1.27 | 1.08 | 1.13 | 1.08 |
| contractile ring contraction involved in cell cycle cytokinesis | 7 | 1.0 | 1.24 | 1.32 | 1.18 | 1.42 | 1.29 | 1.17 |
| cytokinetic cell separation | 1 | -1.24 | -1.05 | 1.11 | 1.18 | 1.4 | -1.36 | -1.18 |
| spindle pole | 23 | 1.28 | 1.12 | 1.43 | 1.06 | 1.39 | 1.41 | 1.26 |
| gamma-tubulin complex | 8 | 1.55 | 1.01 | 1.26 | 1.07 | 1.31 | 1.48 | 1.15 |
| gamma-tubulin large complex | 5 | 1.67 | -1.04 | 1.23 | 1.09 | 1.31 | 1.41 | 1.1 |
| cytoplasmic mRNA processing body | 13 | 1.31 | 1.27 | 1.6 | 1.18 | 1.51 | 1.59 | 1.3 |
| condensed chromosome inner kinetochore | 1 | 1.0 | 1.07 | -1.07 | 1.19 | -1.13 | -1.28 | 1.01 |
| condensed chromosome outer kinetochore | 7 | -1.03 | 1.07 | 1.16 | 1.1 | 1.38 | -1.08 | -1.04 |
| condensed nuclear chromosome inner kinetochore | 1 | 1.0 | 1.07 | -1.07 | 1.19 | -1.13 | -1.28 | 1.01 |
| base pairing with rRNA | 2 | 1.38 | 1.09 | 1.14 | 1.35 | 1.29 | -1.02 | -1.04 |
| nuclear-transcribed mRNA catabolic process | 23 | 1.11 | 1.16 | 1.34 | 1.18 | 1.37 | 1.32 | 1.21 |
| mitochondrial RNA metabolic process | 3 | 1.29 | 1.23 | 1.41 | 1.12 | 1.47 | 1.73 | -1.1 |
| regulatory region DNA binding | 74 | 1.02 | 1.03 | 1.06 | 1.02 | 1.11 | 1.13 | 1.07 |
| transcription regulatory region sequence-specific DNA binding | 65 | 1.03 | 1.01 | 1.06 | 1.01 | 1.09 | 1.15 | 1.08 |
| RNA polymerase II regulatory region sequence-specific DNA binding | 45 | 1.09 | -1.0 | 1.06 | 1.02 | 1.1 | 1.12 | 1.05 |
| RNA polymerase II core promoter proximal region sequence-specific DNA binding | 4 | 1.25 | 1.09 | 1.19 | 1.0 | 1.33 | 1.57 | 1.45 |
| RNA polymerase II distal enhancer sequence-specific DNA binding | 17 | 1.22 | 1.05 | 1.08 | -1.03 | 1.01 | 1.06 | 1.21 |
| sequence-specific DNA binding RNA polymerase II transcription factor activity | 150 | 1.13 | 1.02 | 1.1 | -1.01 | 1.12 | 1.15 | 1.16 |
| RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity | 59 | 1.12 | 1.09 | 1.16 | 1.01 | 1.21 | 1.12 | 1.21 |
| RNA polymerase II core promoter sequence-specific DNA binding transcription factor activity | 19 | 1.4 | 1.13 | 1.24 | 1.2 | 1.34 | 1.2 | 1.07 |
| core promoter proximal region sequence-specific DNA binding | 5 | 1.34 | 1.15 | 1.24 | 1.06 | 1.3 | 1.49 | 1.32 |
| protein binding transcription factor activity | 95 | 1.28 | 1.11 | 1.27 | 1.15 | 1.34 | 1.24 | 1.16 |
| transcription factor binding transcription factor activity | 95 | 1.28 | 1.11 | 1.27 | 1.15 | 1.34 | 1.24 | 1.16 |
| RNA polymerase II regulatory region DNA binding | 47 | 1.09 | 1.0 | 1.06 | 1.02 | 1.1 | 1.12 | 1.04 |
| core promoter sequence-specific DNA binding | 2 | -1.01 | 1.32 | 1.26 | 1.21 | 1.46 | 1.11 | 1.06 |
| core promoter binding | 2 | -1.01 | 1.32 | 1.26 | 1.21 | 1.46 | 1.11 | 1.06 |
| regulatory region nucleic acid binding | 74 | 1.02 | 1.03 | 1.06 | 1.02 | 1.11 | 1.13 | 1.07 |
| nucleic acid binding transcription factor activity | 411 | 1.09 | 1.04 | 1.09 | 1.01 | 1.1 | 1.11 | 1.09 |
| sequence-specific core promoter binding RNA polymerase II transcription factor activity involved in preinitiation complex assembly | 18 | 1.41 | 1.13 | 1.25 | 1.21 | 1.36 | 1.2 | 1.08 |
| RNA polymerase II transcription factor binding transcription factor activity | 46 | 1.44 | 1.04 | 1.23 | 1.18 | 1.35 | 1.26 | 1.1 |
| RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription | 40 | 1.12 | 1.13 | 1.2 | 1.01 | 1.21 | 1.17 | 1.23 |
| RNA polymerase II core promoter proximal region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription | 21 | 1.21 | 1.02 | 1.09 | 1.02 | 1.19 | 1.01 | 1.17 |
| RNA polymerase II basal transcription factor binding transcription factor activity | 6 | 1.58 | 1.1 | 1.46 | 1.16 | 1.41 | 1.29 | 1.2 |
| RNA polymerase II transcription factor binding | 15 | 1.46 | -1.02 | 1.01 | 1.1 | 1.19 | 1.25 | 1.17 |
| RNA polymerase II basal transcription factor binding | 7 | 1.26 | 1.21 | 1.29 | 1.22 | 1.32 | 1.14 | 1.2 |
| TFIIA-class transcription factor binding | 3 | 1.34 | 1.29 | 1.39 | 1.2 | 1.61 | 1.37 | 1.28 |
| TFIIB-class transcription factor binding | 1 | 1.15 | 1.32 | 1.1 | 1.22 | 1.43 | 1.06 | 1.02 |
| TFIID-class transcription factor binding | 2 | 1.34 | 1.01 | -1.02 | 1.28 | 1.04 | -1.14 | -1.12 |
| TFIIF-class transcription factor binding | 2 | 1.1 | 1.32 | 1.52 | 1.2 | 1.24 | 1.14 | 1.45 |
| basal transcription machinery binding | 8 | 1.09 | 1.07 | 1.15 | 1.14 | 1.23 | 1.02 | 1.08 |
| basal RNA polymerase II transcription machinery binding | 8 | 1.09 | 1.07 | 1.15 | 1.14 | 1.23 | 1.02 | 1.08 |
| response to acid | 5 | 1.44 | -1.04 | 1.24 | 1.2 | 1.26 | 1.13 | -1.15 |
| RNA polymerase II activating transcription factor binding | 4 | 2.11 | -1.73 | -1.85 | -1.26 | -1.03 | 1.44 | 1.34 |
| RNA polymerase II repressing transcription factor binding | 2 | 1.24 | 1.19 | 1.36 | 1.24 | 1.29 | 1.42 | 1.1 |
| RNA polymerase II transcription cofactor activity | 41 | 1.42 | 1.04 | 1.22 | 1.19 | 1.34 | 1.27 | 1.09 |
| RNA polymerase II transcription coactivator activity | 8 | 1.57 | -1.19 | -1.01 | 1.03 | 1.27 | 1.49 | 1.13 |
| RNA polymerase II transcription corepressor activity | 2 | 1.12 | 1.2 | 1.26 | 1.29 | 1.7 | 1.19 | 1.25 |
| promoter clearance during DNA-dependent transcription | 5 | 1.21 | 1.11 | 1.42 | 1.33 | 1.33 | 1.15 | 1.09 |
| promoter clearance from RNA polymerase II promoter | 5 | 1.21 | 1.11 | 1.42 | 1.33 | 1.33 | 1.15 | 1.09 |
| DNA-dependent transcriptional open complex formation | 5 | 1.21 | 1.11 | 1.42 | 1.33 | 1.33 | 1.15 | 1.09 |
| transcriptional open complex formation at RNA polymerase II promoter | 5 | 1.21 | 1.11 | 1.42 | 1.33 | 1.33 | 1.15 | 1.09 |
| protein-DNA complex remodeling | 5 | 1.21 | 1.11 | 1.42 | 1.33 | 1.33 | 1.15 | 1.09 |
| RNA polymerase II transcription coactivator activity involved in preinitiation complex assembly | 2 | 1.66 | -1.04 | 1.36 | 1.21 | 1.19 | 1.16 | -1.0 |
| TBP-class protein binding RNA polymerase II transcription factor activity involved in preinitiation complex assembly | 6 | 1.58 | 1.1 | 1.46 | 1.16 | 1.41 | 1.29 | 1.2 |
| TBP-class protein binding RNA polymerase II transcription factor activity | 6 | 1.58 | 1.1 | 1.46 | 1.16 | 1.41 | 1.29 | 1.2 |
| enhancer sequence-specific DNA binding | 18 | 1.2 | 1.05 | 1.07 | -1.02 | 1.0 | 1.04 | 1.19 |
| core promoter proximal region DNA binding | 5 | 1.34 | 1.15 | 1.24 | 1.06 | 1.3 | 1.49 | 1.32 |
| DNA-dependent transcriptional start site selection | 1 | 2.19 | 1.23 | -1.08 | 1.33 | 1.17 | -1.1 | 1.1 |
| transcriptional start site selection at RNA polymerase II promoter | 1 | 2.19 | 1.23 | -1.08 | 1.33 | 1.17 | -1.1 | 1.1 |
| RNA polymerase II transcription factor binding transcription factor activity involved in positive regulation of transcription | 8 | 1.57 | -1.19 | -1.01 | 1.03 | 1.27 | 1.49 | 1.13 |
| RNA polymerase II transcription factor binding transcription factor activity involved in negative regulation of transcription | 3 | 1.37 | 1.05 | 1.07 | 1.1 | 1.48 | -1.12 | 1.13 |
| age-dependent response to oxidative stress | 1 | -1.1 | 1.09 | 1.19 | 1.01 | 1.01 | 1.19 | 1.25 |
| entry into host | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| neurotransmitter uptake | 1 | -1.06 | -1.11 | -1.0 | -1.08 | -1.15 | 3.37 | -1.04 |
| regulation of neurotransmitter levels | 106 | -1.07 | 1.06 | 1.19 | 1.11 | 1.22 | 1.06 | 1.07 |
| neurotransmitter biosynthetic process | 4 | 1.15 | -1.01 | -1.13 | -1.06 | 1.01 | -1.12 | -1.05 |
| acetylcholine catabolic process in synaptic cleft | 1 | 2.27 | 1.23 | -5.17 | -1.59 | -2.2 | -9.9 | 9.42 |
| regulation of action potential | 8 | 1.57 | -1.04 | 1.3 | -1.12 | 1.17 | 2.65 | 1.29 |
| cysteine-type endopeptidase activity | 29 | -1.39 | -1.19 | 1.01 | -1.08 | 1.27 | -1.29 | -1.2 |
| RNA methylation | 10 | 1.62 | 1.18 | 1.23 | 1.26 | 1.35 | 1.29 | 1.02 |
| microfibril | 1 | 1.02 | -1.0 | 1.14 | 1.18 | 1.1 | 1.03 | -1.08 |
| calcium ion binding | 192 | 1.36 | -1.02 | 1.1 | -1.05 | 1.09 | 1.28 | 1.2 |
| selenocysteine incorporation | 1 | 1.74 | 1.02 | 1.02 | 1.41 | 1.48 | -1.49 | -1.06 |
| voltage-gated sodium channel complex | 1 | 1.49 | 1.1 | 1.18 | 1.37 | 1.22 | 1.19 | 1.04 |
| peptide amidation | 1 | 2.74 | 1.3 | 1.75 | 1.08 | 1.98 | 1.97 | 2.28 |
| pseudouridine synthesis | 12 | 1.48 | 1.3 | 1.18 | 1.15 | 1.18 | 1.37 | 1.11 |
| retinoid metabolic process | 3 | 1.01 | -1.01 | 1.02 | 1.05 | -1.08 | 1.07 | -1.1 |
| oxygen transporter activity | 11 | 1.36 | 1.04 | 1.04 | -1.04 | -1.15 | 1.01 | -1.01 |
| proteoglycan metabolic process | 18 | 1.24 | 1.14 | 1.2 | 1.07 | 1.15 | 1.23 | 1.5 |
| sulfur compound metabolic process | 37 | -1.0 | 1.04 | 1.02 | 1.1 | 1.02 | 1.18 | 1.06 |
| lipopolysaccharide binding | 1 | 8.83 | -1.7 | -1.73 | -1.63 | -1.34 | 3.89 | 1.3 |
| ciliary or flagellar motility | 2 | 1.0 | -1.02 | -1.01 | -1.04 | -1.08 | -1.04 | 1.1 |
| beta-amyloid binding | 1 | -1.11 | 1.12 | -1.05 | 1.06 | -1.09 | -1.06 | -1.01 |
| ovarian follicle development | 1 | -1.49 | 2.58 | -1.26 | -1.23 | -1.23 | -1.38 | 1.24 |
| oocyte growth | 2 | 1.25 | 1.36 | 1.54 | 1.22 | 1.6 | 1.07 | 1.72 |
| oocyte maturation | 3 | -1.0 | 1.13 | -1.07 | -1.01 | -1.03 | -1.0 | 1.1 |
| metabolic process | 3907 | -1.04 | -1.03 | -1.04 | 1.02 | 1.06 | -1.04 | -1.05 |
| cell growth | 59 | 1.22 | 1.13 | 1.38 | 1.04 | 1.43 | 1.35 | 1.28 |
| regulation of cell growth | 38 | 1.09 | 1.19 | 1.46 | 1.1 | 1.43 | 1.53 | 1.25 |
| heterophilic cell-cell adhesion | 12 | -1.44 | 1.04 | 1.3 | -1.02 | 1.46 | -1.24 | 1.32 |
| galactoside binding | 2 | 1.45 | -1.07 | 1.88 | 1.03 | 1.06 | 1.31 | 1.14 |
| microtubule bundle formation | 16 | -1.31 | 1.1 | 1.24 | 1.11 | 1.2 | -1.29 | -1.07 |
| detection of chemical stimulus involved in sensory perception of bitter taste | 1 | -1.77 | -1.51 | -1.46 | -1.66 | 9.03 | 1.38 | -1.43 |
| detection of chemical stimulus involved in sensory perception of sweet taste | 1 | 15.43 | 1.05 | -1.1 | -1.34 | 2.06 | 11.73 | 3.15 |
| Gi/o-coupled serotonin receptor activity | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| Gq/11-coupled serotonin receptor activity | 1 | 1.13 | -1.11 | -1.04 | -1.12 | -1.18 | 1.14 | 1.05 |
| dopamine receptor activity, coupled via Gs | 2 | -1.21 | 1.11 | -1.21 | 1.0 | -1.21 | -1.22 | -1.28 |
| dopamine receptor activity, coupled via Gi/Go | 1 | -1.08 | -1.06 | -1.29 | 1.12 | -1.04 | -1.27 | -1.07 |
| neuromedin U receptor activity | 1 | -1.22 | -1.11 | -1.12 | -1.18 | -1.25 | -1.3 | -1.2 |
| G-protein coupled adenosine receptor activity | 1 | -1.32 | 1.16 | 1.21 | -1.14 | -1.21 | 1.47 | 1.08 |
| dense fibrillar component | 1 | -1.19 | 1.16 | -1.05 | 1.68 | 1.27 | -1.6 | -1.28 |
| peptide receptor activity | 45 | 1.11 | -1.06 | 1.03 | -1.06 | -1.05 | 1.15 | 1.01 |
| eye development | 265 | 1.14 | 1.1 | 1.26 | 1.06 | 1.27 | 1.23 | 1.24 |
| urogenital system development | 69 | 1.2 | -1.03 | 1.12 | -1.01 | 1.21 | 1.35 | 1.08 |
| conditioned taste aversion | 3 | 1.58 | 1.48 | 1.62 | -1.13 | 1.26 | 1.28 | 1.57 |
| G-protein coupled receptor binding | 7 | -1.1 | -1.01 | 1.12 | -1.08 | 1.17 | 1.16 | 1.2 |
| response to hypoxia | 43 | 1.14 | 1.14 | 1.22 | 1.08 | 1.27 | 1.28 | 1.2 |
| ameboidal cell migration | 35 | 1.62 | -1.01 | 1.31 | -1.06 | 1.21 | 1.46 | 1.11 |
| acrosomal vesicle | 1 | -1.22 | 1.14 | 1.06 | 1.01 | -1.52 | -1.37 | 1.04 |
| ATPase activator activity | 3 | 1.77 | 1.07 | 1.2 | 1.24 | 1.37 | 1.32 | 1.35 |
| regulation of chromatin assembly or disassembly | 6 | 1.1 | 1.2 | 1.31 | 1.12 | 1.3 | 1.19 | 1.21 |
| male germ cell nucleus | 10 | -1.1 | 1.13 | 1.01 | 1.03 | -1.05 | -1.24 | 1.05 |
| female germ cell nucleus | 1 | 1.04 | 1.22 | 1.16 | 1.15 | 1.04 | -1.0 | -1.09 |
| long-chain fatty acid metabolic process | 8 | 2.31 | -2.32 | -1.73 | -1.98 | -1.14 | -1.49 | 1.69 |
| formation of translation initiation ternary complex | 1 | 1.02 | 1.21 | 1.26 | 1.24 | 1.31 | 1.17 | 1.22 |
| tRNA 5'-leader removal | 1 | -1.05 | 1.03 | -1.13 | -1.0 | -1.0 | -1.23 | -1.11 |
| axonemal dynein complex | 12 | -1.02 | 1.03 | 1.04 | 1.02 | -1.05 | 1.0 | -1.11 |
| cytoplasmic dynein complex | 11 | -1.01 | 1.07 | 1.14 | 1.01 | 1.06 | 1.15 | -1.02 |
| histamine metabolic process | 2 | 1.02 | -1.27 | 1.33 | 1.09 | 1.3 | 1.8 | 1.14 |
| histamine biosynthetic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| embryonic development via the syncytial blastoderm | 159 | 1.1 | 1.06 | 1.3 | 1.03 | 1.34 | 1.33 | 1.22 |
| gastrulation with mouth forming first | 30 | 1.11 | -1.0 | 1.2 | 1.03 | 1.32 | 1.16 | 1.06 |
| formation of primary germ layer | 28 | 1.53 | -1.01 | 1.18 | -1.09 | 1.11 | 1.67 | 1.15 |
| ectoderm formation | 2 | 4.98 | -1.19 | 1.75 | -1.46 | 1.16 | 1.31 | 1.35 |
| endoderm formation | 4 | 2.13 | -1.15 | 1.08 | -1.19 | 1.07 | 2.11 | 1.76 |
| mesoderm formation | 26 | 1.41 | 1.01 | 1.15 | -1.07 | 1.1 | 1.6 | 1.11 |
| cell fate specification | 59 | 1.11 | 1.06 | 1.1 | 1.04 | 1.17 | -1.02 | 1.22 |
| cell fate determination | 122 | 1.2 | 1.07 | 1.23 | 1.02 | 1.24 | 1.19 | 1.2 |
| mesodermal cell fate commitment | 16 | 1.47 | 1.02 | 1.02 | -1.04 | 1.04 | 1.32 | 1.13 |
| endodermal cell fate commitment | 1 | 8.12 | -1.58 | 1.44 | -1.54 | 1.01 | 1.69 | 1.21 |
| ectodermal cell fate commitment | 2 | 4.98 | -1.19 | 1.75 | -1.46 | 1.16 | 1.31 | 1.35 |
| ectodermal cell fate specification | 2 | 4.98 | -1.19 | 1.75 | -1.46 | 1.16 | 1.31 | 1.35 |
| intermediate filament | 2 | 1.28 | 1.06 | -1.0 | 1.32 | 1.15 | -1.17 | -1.1 |
| ruffle | 2 | 1.28 | 1.39 | 2.24 | -1.02 | 2.03 | 1.81 | 1.72 |
| lipid kinase activity | 2 | 1.57 | 1.08 | 1.68 | -1.01 | 1.59 | 1.77 | 1.96 |
| ceramide kinase activity | 1 | 2.79 | -1.02 | 1.66 | -1.11 | 1.24 | 2.25 | 2.21 |
| establishment of planar polarity | 70 | 1.2 | 1.06 | 1.23 | 1.01 | 1.28 | 1.24 | 1.46 |
| establishment of imaginal disc-derived wing hair orientation | 22 | 1.56 | 1.08 | 1.28 | 1.02 | 1.4 | 1.32 | 1.3 |
| morphogenesis of a polarized epithelium | 79 | 1.19 | 1.08 | 1.28 | 1.02 | 1.3 | 1.27 | 1.44 |
| oenocyte differentiation | 8 | 1.23 | 1.09 | 1.12 | 1.12 | 1.24 | 1.37 | 1.16 |
| optic placode formation | 4 | -1.14 | 1.1 | 1.23 | 1.12 | 1.47 | -1.2 | -1.08 |
| optic lobe placode formation | 4 | -1.14 | 1.1 | 1.23 | 1.12 | 1.47 | -1.2 | -1.08 |
| compound eye morphogenesis | 198 | 1.16 | 1.1 | 1.25 | 1.06 | 1.25 | 1.2 | 1.26 |
| Bolwig's organ morphogenesis | 9 | 1.23 | -1.01 | 1.15 | 1.06 | 1.23 | 1.18 | 1.03 |
| optic lobe placode development | 8 | -1.18 | 1.11 | 1.22 | 1.1 | 1.34 | -1.02 | 1.03 |
| compound eye photoreceptor cell differentiation | 117 | 1.22 | 1.11 | 1.3 | 1.08 | 1.29 | 1.25 | 1.2 |
| compound eye photoreceptor fate commitment | 51 | 1.32 | 1.09 | 1.23 | 1.09 | 1.32 | 1.24 | 1.18 |
| compound eye photoreceptor development | 60 | 1.11 | 1.12 | 1.33 | 1.04 | 1.24 | 1.21 | 1.22 |
| eye photoreceptor cell differentiation | 120 | 1.21 | 1.12 | 1.32 | 1.08 | 1.29 | 1.26 | 1.21 |
| morphogenesis of a branching structure | 48 | 1.11 | 1.11 | 1.43 | 1.03 | 1.35 | 1.27 | 1.3 |
| neuron migration | 1 | -1.42 | 1.01 | 1.2 | 1.29 | 2.44 | -1.59 | 1.35 |
| cell activation | 10 | 1.19 | 1.04 | 1.07 | 1.06 | -1.05 | 1.15 | 1.06 |
| phosphotyrosine binding | 1 | -1.42 | 1.22 | 1.22 | 1.19 | 1.41 | -1.04 | -1.04 |
| kidney development | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| epithelial to mesenchymal transition | 1 | 1.47 | 1.13 | -1.02 | 1.1 | -1.07 | -1.03 | 1.1 |
| embryonic epithelial tube formation | 6 | 1.03 | -1.09 | 1.01 | -1.18 | -1.08 | 1.45 | 1.48 |
| opsonin receptor activity | 1 | 1.18 | 1.16 | 1.11 | 1.22 | 1.12 | 1.2 | 1.16 |
| pattern binding | 116 | -1.88 | -1.54 | -1.66 | -1.39 | -1.53 | -1.88 | 1.19 |
| response to yeast | 1 | 5.18 | -1.34 | 1.33 | -1.5 | 1.33 | 6.7 | 1.77 |
| nucleoside binding | 5 | -2.97 | 1.34 | -1.45 | 1.07 | -1.17 | -2.3 | -1.24 |
| endothelial cell development | 3 | 1.7 | 1.01 | 1.41 | -1.21 | 1.32 | 4.52 | 2.14 |
| selenium compound metabolic process | 2 | 1.76 | 1.25 | 1.23 | 1.04 | 1.04 | 1.62 | -1.07 |
| glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity | 1 | 1.03 | 1.3 | 1.31 | 1.09 | 1.28 | -1.03 | 1.49 |
| tissue homeostasis | 20 | 1.33 | -1.02 | 1.13 | -1.12 | 1.22 | 1.46 | 1.54 |
| retina homeostasis | 2 | -1.1 | 1.15 | 1.07 | 1.1 | 1.01 | 1.07 | 1.17 |
| regulation of protein phosphorylation | 65 | 1.23 | 1.16 | 1.37 | 1.12 | 1.3 | 1.37 | 1.18 |
| negative regulation of protein phosphorylation | 2 | -1.04 | 1.11 | 1.79 | -1.0 | 1.29 | 1.24 | 1.01 |
| positive regulation of protein phosphorylation | 5 | 1.08 | 1.17 | 1.78 | 1.11 | 1.35 | 1.9 | 1.34 |
| regulation of cell-matrix adhesion | 2 | 1.91 | 1.0 | 2.08 | 1.07 | 1.96 | 1.63 | 1.31 |
| negative regulation of cell-matrix adhesion | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| startle response | 13 | 1.09 | 1.15 | 1.46 | 1.07 | 1.52 | 1.19 | 1.26 |
| G-protein alpha-subunit binding | 2 | -1.16 | 1.18 | -1.02 | 1.16 | 1.01 | -1.6 | -1.07 |
| morphogenesis of an epithelium | 238 | 1.18 | 1.05 | 1.28 | 1.02 | 1.3 | 1.33 | 1.27 |
| morphogenesis of an epithelial sheet | 1 | 1.57 | 1.36 | 1.53 | 1.09 | 1.59 | 1.28 | 1.35 |
| regulation of heart rate | 3 | -2.33 | 1.98 | 1.4 | -1.73 | -1.39 | 2.29 | -1.17 |
| regulation of sodium ion transport | 6 | -1.09 | 1.11 | 1.21 | 1.02 | 1.05 | -1.15 | 1.01 |
| p53 binding | 1 | 1.78 | 1.16 | 2.67 | 1.12 | 2.53 | 3.19 | 2.15 |
| positive regulation of neuroblast proliferation | 2 | -1.35 | 1.28 | 1.19 | 1.29 | 1.21 | -1.02 | 1.09 |
| epithelial cell development | 14 | 1.16 | 1.1 | 1.33 | 1.01 | 1.23 | 1.71 | 1.39 |
| protein depalmitoylation | 2 | 2.75 | 1.12 | 1.29 | 1.19 | 1.61 | 1.16 | 1.22 |
| regulation of receptor internalization | 2 | 2.64 | -1.34 | 1.04 | -1.28 | -1.11 | 2.63 | 1.36 |
| positive regulation of receptor internalization | 2 | 2.64 | -1.34 | 1.04 | -1.28 | -1.11 | 2.63 | 1.36 |
| polyprenyltransferase activity | 1 | 1.04 | 1.26 | 1.1 | 1.28 | -1.02 | -1.97 | -1.12 |
| tRNA wobble base modification | 2 | 1.94 | 1.19 | 1.23 | 1.17 | 1.27 | 1.92 | 1.12 |
| tRNA wobble uridine modification | 2 | 1.94 | 1.19 | 1.23 | 1.17 | 1.27 | 1.92 | 1.12 |
| podosome | 1 | -1.23 | -1.76 | -1.82 | -1.76 | -2.16 | 17.9 | -1.88 |
| store-operated calcium entry | 1 | 2.66 | -1.14 | 1.6 | 1.07 | 1.44 | 4.2 | 2.14 |
| aggressive behavior | 44 | 1.19 | 1.05 | 1.23 | -1.02 | 1.15 | 1.61 | 1.24 |
| inter-male aggressive behavior | 42 | 1.18 | 1.06 | 1.2 | 1.0 | 1.15 | 1.58 | 1.25 |
| G-quadruplex RNA binding | 1 | 1.1 | 1.52 | 1.31 | 1.18 | 1.34 | 1.42 | 1.38 |
| larval development | 73 | 1.16 | 1.08 | 1.33 | 1.02 | 1.21 | 1.2 | 1.27 |
| instar larval or pupal development | 424 | 1.18 | 1.05 | 1.22 | 1.02 | 1.26 | 1.27 | 1.22 |
| instar larval development | 48 | 1.09 | 1.08 | 1.31 | 1.01 | 1.22 | 1.22 | 1.26 |
| palmitoyltransferase complex | 1 | -1.92 | 1.11 | 2.34 | 1.27 | 1.64 | -1.44 | -1.26 |
| defense response to insect | 1 | 23.01 | -3.14 | -2.9 | -3.17 | -2.48 | 1.73 | 2.58 |
| positive regulation of antimicrobial peptide production | 1 | -1.72 | 1.19 | -1.18 | 1.31 | 1.64 | 3.16 | 1.11 |
| innate immune response in mucosa | 1 | -1.37 | -1.13 | -1.19 | -1.54 | 1.82 | 7.35 | 2.8 |
| response to molecule of bacterial origin | 3 | 2.15 | 1.03 | 1.06 | -1.04 | 1.33 | 1.63 | 1.06 |
| adaptive immune response | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| organ or tissue specific immune response | 7 | 1.24 | 1.06 | 1.29 | -1.06 | 1.56 | 1.62 | 1.43 |
| immune effector process | 20 | 1.79 | -1.15 | 1.28 | -1.11 | 1.26 | 1.41 | 1.26 |
| activation of immune response | 1 | 1.17 | 1.46 | 1.63 | 1.36 | 1.42 | 1.78 | 1.19 |
| myeloid leukocyte activation | 1 | 2.18 | -2.11 | 2.19 | 1.49 | 1.48 | 1.37 | 1.38 |
| immune system process | 193 | 1.3 | -1.15 | -1.02 | -1.12 | 1.05 | 1.17 | 1.2 |
| mucosal immune response | 7 | 1.24 | 1.06 | 1.29 | -1.06 | 1.56 | 1.62 | 1.43 |
| immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 3 | 1.28 | -1.17 | 2.05 | 1.18 | 1.64 | 1.24 | 1.18 |
| inflammatory response to antigenic stimulus | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| acute inflammatory response to antigenic stimulus | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| production of molecular mediator of immune response | 2 | -1.54 | 1.11 | 1.81 | 1.12 | 1.7 | 3.17 | 1.43 |
| leukocyte mediated immunity | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| immune system development | 52 | 1.16 | 1.07 | 1.33 | 1.05 | 1.3 | 1.2 | 1.34 |
| hypersensitivity | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| acute inflammatory response | 3 | -1.45 | 1.14 | 1.29 | 1.15 | 1.43 | -1.36 | 1.17 |
| regulation of immune system process | 73 | 1.33 | -1.01 | 1.08 | -1.01 | 1.11 | 1.27 | 1.26 |
| negative regulation of immune system process | 16 | 1.45 | 1.01 | 1.09 | -1.01 | 1.17 | 1.27 | 1.37 |
| positive regulation of immune system process | 8 | 1.43 | 1.03 | 1.15 | 1.01 | 1.19 | 1.8 | 1.48 |
| regulation of immune effector process | 36 | 1.47 | -1.08 | 1.0 | -1.04 | 1.04 | 1.31 | 1.19 |
| positive regulation of immune effector process | 1 | -1.72 | 1.19 | -1.18 | 1.31 | 1.64 | 3.16 | 1.11 |
| regulation of production of molecular mediator of immune response | 34 | 1.45 | -1.09 | -1.02 | -1.05 | 1.03 | 1.32 | 1.2 |
| positive regulation of production of molecular mediator of immune response | 1 | -1.72 | 1.19 | -1.18 | 1.31 | 1.64 | 3.16 | 1.11 |
| regulation of antimicrobial humoral response | 39 | 1.26 | -1.07 | 1.01 | -1.04 | 1.05 | 1.16 | 1.17 |
| antimicrobial peptide production | 2 | -1.54 | 1.11 | 1.81 | 1.12 | 1.7 | 3.17 | 1.43 |
| antifungal peptide production | 1 | -1.72 | 1.19 | -1.18 | 1.31 | 1.64 | 3.16 | 1.11 |
| regulation of antimicrobial peptide production | 34 | 1.45 | -1.09 | -1.02 | -1.05 | 1.03 | 1.32 | 1.2 |
| regulation of antibacterial peptide production | 19 | 1.47 | -1.12 | 1.02 | -1.12 | 1.06 | 1.26 | 1.29 |
| regulation of antifungal peptide production | 7 | 2.0 | -1.32 | -1.17 | 1.03 | 1.03 | 1.92 | 1.18 |
| peptide secretion | 1 | 1.36 | -1.09 | 1.47 | 1.03 | 1.67 | -1.23 | 2.47 |
| regulation of peptide secretion | 1 | 1.12 | 1.15 | 1.24 | 1.0 | 1.0 | 1.1 | -1.06 |
| positive regulation of peptide secretion | 1 | 1.12 | 1.15 | 1.24 | 1.0 | 1.0 | 1.1 | -1.06 |
| positive regulation of antifungal peptide production | 1 | -1.72 | 1.19 | -1.18 | 1.31 | 1.64 | 3.16 | 1.11 |
| regulation of antimicrobial peptide biosynthetic process | 34 | 1.45 | -1.09 | -1.02 | -1.05 | 1.03 | 1.32 | 1.2 |
| negative regulation of antimicrobial peptide biosynthetic process | 2 | 1.03 | 1.14 | 1.57 | 1.07 | 1.57 | -1.03 | -1.32 |
| positive regulation of antimicrobial peptide biosynthetic process | 23 | 1.75 | -1.22 | -1.09 | -1.11 | -1.01 | 1.39 | 1.29 |
| regulation of antibacterial peptide biosynthetic process | 19 | 1.47 | -1.12 | 1.02 | -1.12 | 1.06 | 1.26 | 1.29 |
| negative regulation of antibacterial peptide biosynthetic process | 2 | 1.03 | 1.14 | 1.57 | 1.07 | 1.57 | -1.03 | -1.32 |
| regulation of antifungal peptide biosynthetic process | 6 | 2.45 | -1.42 | -1.16 | -1.01 | -1.05 | 1.77 | 1.2 |
| regulation of biosynthetic process of antibacterial peptides active against Gram-negative bacteria | 7 | 1.96 | -1.11 | 1.11 | -1.19 | 1.09 | 1.65 | 1.22 |
| negative regulation of biosynthetic process of antibacterial peptides active against Gram-negative bacteria | 2 | 1.03 | 1.14 | 1.57 | 1.07 | 1.57 | -1.03 | -1.32 |
| regulation of biosynthetic process of antibacterial peptides active against Gram-positive bacteria | 4 | 1.05 | -1.9 | -2.24 | -1.85 | -1.9 | -1.18 | 1.68 |
| regulation of response to biotic stimulus | 44 | 1.36 | -1.07 | 1.05 | -1.02 | 1.04 | 1.11 | 1.1 |
| negative regulation of response to biotic stimulus | 7 | 1.37 | 1.0 | 1.62 | 1.21 | 1.38 | -1.12 | -1.13 |
| positive regulation of response to biotic stimulus | 1 | 8.35 | -1.25 | -1.3 | -1.07 | -1.15 | 1.62 | -1.07 |
| regulation of humoral immune response | 39 | 1.26 | -1.07 | 1.01 | -1.04 | 1.05 | 1.16 | 1.17 |
| negative regulation of humoral immune response | 4 | 1.35 | 1.1 | 1.46 | 1.15 | 1.8 | 1.12 | 1.2 |
| generation of a signal involved in cell-cell signaling | 101 | -1.07 | 1.04 | 1.24 | 1.13 | 1.26 | 1.1 | 1.03 |
| regionalization | 378 | 1.18 | 1.05 | 1.18 | 1.02 | 1.22 | 1.26 | 1.24 |
| developmental process involved in reproduction | 482 | 1.13 | 1.1 | 1.22 | 1.05 | 1.26 | 1.18 | 1.17 |
| heart morphogenesis | 12 | 1.02 | -1.01 | 1.12 | 1.1 | 1.36 | 1.23 | 1.2 |
| system process | 634 | 1.08 | 1.03 | 1.09 | -1.0 | 1.08 | 1.15 | 1.06 |
| skeletal muscle contraction | 2 | -1.89 | -1.52 | -1.42 | -1.43 | -1.62 | -1.61 | 2.67 |
| voluntary skeletal muscle contraction | 1 | -1.18 | -1.0 | 1.06 | -1.02 | -1.12 | 1.04 | 1.04 |
| muscle system process | 13 | 1.35 | -1.07 | 1.09 | -1.24 | 1.12 | 1.46 | 1.24 |
| circulatory system process | 8 | 1.03 | 1.14 | -1.1 | 1.05 | 1.17 | 1.15 | 1.14 |
| renal system process | 6 | -4.46 | 1.18 | -1.92 | -1.14 | -3.52 | -1.44 | -1.93 |
| heart process | 8 | 1.03 | 1.14 | -1.1 | 1.05 | 1.17 | 1.15 | 1.14 |
| detection of carbon dioxide | 2 | -1.18 | -1.02 | 1.02 | -1.08 | -1.1 | -1.05 | -1.18 |
| negative regulation of heart induction by canonical Wnt receptor signaling pathway | 3 | 1.09 | 1.1 | 1.13 | 1.08 | 1.18 | 1.14 | 2.09 |
| outflow tract morphogenesis | 3 | -1.03 | 1.08 | 1.55 | 1.17 | 2.15 | 1.03 | 1.48 |
| regulation of organ formation | 8 | 1.56 | -1.2 | -1.1 | 1.04 | 1.14 | 1.2 | -1.02 |
| endothelium development | 3 | 1.7 | 1.01 | 1.41 | -1.21 | 1.32 | 4.52 | 2.14 |
| Wnt receptor signaling pathway involved in heart development | 3 | 1.09 | 1.1 | 1.13 | 1.08 | 1.18 | 1.14 | 2.09 |
| amino acid transmembrane transport | 30 | -1.61 | -1.12 | -1.51 | -1.16 | -1.02 | -1.08 | -1.1 |
| epithelial cell morphogenesis | 9 | 1.12 | 1.09 | 1.33 | 1.05 | 1.18 | 1.38 | 1.22 |
| apical constriction | 4 | -1.26 | 1.28 | 1.59 | 1.06 | 1.39 | 1.15 | 1.23 |
| cytoneme morphogenesis | 2 | -1.85 | -1.12 | 1.68 | 1.29 | 1.66 | 1.71 | -2.53 |
| neural retina development | 1 | -1.1 | -1.15 | -1.07 | -1.18 | 1.52 | 1.28 | 1.24 |
| nucleic acid binding | 1407 | 1.18 | 1.11 | 1.19 | 1.1 | 1.2 | 1.16 | 1.11 |
| DNA binding | 669 | 1.15 | 1.09 | 1.17 | 1.08 | 1.17 | 1.12 | 1.11 |
| DNA helicase activity | 34 | 1.08 | 1.15 | 1.32 | 1.19 | 1.42 | 1.11 | 1.09 |
| AT DNA binding | 5 | 1.49 | -1.14 | -1.13 | 1.14 | 1.47 | 1.04 | 1.12 |
| bent DNA binding | 2 | 1.7 | 1.14 | 1.37 | 1.1 | 1.36 | 1.18 | 1.22 |
| chromatin binding | 72 | 1.18 | 1.18 | 1.3 | 1.16 | 1.38 | 1.26 | 1.23 |
| lamin binding | 1 | 1.4 | 1.14 | 1.18 | 1.58 | 1.38 | 1.2 | 1.19 |
| damaged DNA binding | 15 | 1.32 | 1.12 | 1.34 | 1.15 | 1.3 | 1.19 | 1.11 |
| DNA clamp loader activity | 3 | 1.23 | 1.05 | 1.23 | 1.24 | 1.24 | 1.1 | -1.01 |
| double-stranded DNA binding | 10 | 1.24 | 1.11 | 1.11 | 1.14 | 1.22 | 1.09 | -1.07 |
| satellite DNA binding | 5 | 1.25 | 1.13 | 1.51 | 1.21 | 1.5 | 1.17 | 1.18 |
| single-stranded DNA binding | 13 | -1.04 | 1.24 | 1.4 | 1.15 | 1.2 | 1.2 | 1.1 |
| sequence-specific DNA binding transcription factor activity | 411 | 1.09 | 1.04 | 1.09 | 1.01 | 1.1 | 1.11 | 1.09 |
| transcription from RNA polymerase I promoter | 6 | 1.31 | 1.13 | 1.31 | 1.22 | 1.28 | 1.23 | 1.04 |
| transcription from RNA polymerase II promoter | 91 | 1.33 | 1.12 | 1.23 | 1.17 | 1.25 | 1.17 | 1.11 |
| copper ion transport | 7 | -1.13 | 1.18 | 1.31 | 1.05 | -1.07 | 1.09 | 1.04 |
| sequence-specific distal enhancer binding RNA polymerase II transcription factor activity | 50 | 1.18 | -1.11 | -1.01 | -1.13 | -1.1 | 1.13 | 1.1 |
| ligand-activated sequence-specific DNA binding RNA polymerase II transcription factor activity | 20 | 1.3 | 1.06 | 1.02 | -1.03 | 1.08 | 1.23 | 1.26 |
| steroid hormone receptor activity | 19 | 1.11 | 1.08 | 1.02 | -1.0 | 1.09 | 1.16 | 1.16 |
| retinoic acid receptor activity | 1 | 1.03 | 1.25 | 1.07 | 1.02 | 1.08 | 1.24 | 1.08 |
| transcription from RNA polymerase III promoter | 12 | 1.28 | 1.21 | 1.25 | 1.18 | 1.32 | 1.03 | 1.12 |
| regulation of transcription elongation, DNA-dependent | 18 | -1.27 | 1.2 | 1.28 | -1.03 | -1.04 | -1.12 | 1.41 |
| transcription cofactor activity | 90 | 1.26 | 1.11 | 1.26 | 1.16 | 1.34 | 1.25 | 1.15 |
| transcription coactivator activity | 41 | 1.32 | 1.06 | 1.24 | 1.13 | 1.34 | 1.33 | 1.18 |
| transcription corepressor activity | 19 | 1.19 | 1.16 | 1.26 | 1.18 | 1.37 | 1.12 | 1.16 |
| transcription termination, DNA-dependent | 2 | -1.11 | 1.05 | 1.39 | 1.1 | 1.37 | 1.26 | -1.23 |
| termination of RNA polymerase II transcription | 1 | -1.51 | -1.02 | 1.38 | 1.04 | 1.42 | 1.11 | -1.1 |
| cytoplasmic sequestering of transcription factor | 4 | 2.74 | 1.08 | 1.33 | 1.02 | 1.2 | 2.19 | 1.75 |
| RNA binding | 381 | 1.26 | 1.16 | 1.26 | 1.13 | 1.22 | 1.27 | 1.13 |
| RNA helicase activity | 45 | 1.22 | 1.23 | 1.32 | 1.18 | 1.31 | 1.38 | 1.21 |
| double-stranded RNA binding | 16 | 1.33 | -1.04 | 1.19 | -1.13 | 1.16 | 1.42 | 1.51 |
| double-stranded RNA adenosine deaminase activity | 1 | 1.13 | 1.14 | 1.55 | 1.01 | 1.4 | 2.0 | 1.26 |
| single-stranded RNA binding | 20 | 1.32 | 1.14 | 1.42 | 1.11 | 1.32 | 1.44 | 1.23 |
| mRNA binding | 170 | 1.24 | 1.17 | 1.28 | 1.15 | 1.25 | 1.26 | 1.16 |
| mRNA 3'-UTR binding | 17 | 1.34 | 1.14 | 1.46 | 1.02 | 1.31 | 1.89 | 1.29 |
| ribonucleoprotein complex | 427 | 1.18 | 1.1 | 1.2 | 1.16 | 1.24 | 1.21 | -1.02 |
| small nuclear ribonucleoprotein complex | 54 | 1.14 | 1.14 | 1.25 | 1.27 | 1.31 | 1.06 | -1.02 |
| structural constituent of ribosome | 167 | 1.21 | 1.04 | 1.14 | 1.12 | 1.17 | 1.28 | -1.18 |
| translation initiation factor activity | 53 | 1.33 | 1.18 | 1.26 | 1.09 | 1.18 | 1.35 | 1.16 |
| translation elongation factor activity | 16 | 1.48 | 1.11 | 1.26 | 1.06 | 1.23 | 1.46 | 1.05 |
| translation release factor activity | 7 | 1.27 | 1.16 | 1.25 | 1.13 | 1.18 | 1.49 | 1.09 |
| regulation of cell cycle | 240 | 1.19 | 1.13 | 1.28 | 1.13 | 1.34 | 1.28 | 1.16 |
| protein folding | 118 | 1.15 | 1.04 | 1.09 | 1.13 | 1.17 | 1.02 | 1.01 |
| peptidyl-prolyl cis-trans isomerase activity | 26 | 1.38 | 1.03 | 1.1 | 1.15 | 1.25 | 1.1 | 1.02 |
| protein disulfide isomerase activity | 10 | 1.1 | -1.01 | 1.22 | 1.11 | 1.12 | 1.08 | 1.12 |
| chromatin assembly or disassembly | 34 | 1.01 | 1.07 | 1.29 | 1.13 | 1.25 | -1.04 | 1.07 |
| nucleosome assembly | 27 | 1.02 | 1.05 | 1.3 | 1.15 | 1.22 | -1.07 | 1.03 |
| nucleosome organization | 36 | 1.09 | 1.07 | 1.28 | 1.16 | 1.21 | 1.01 | 1.03 |
| histone binding | 10 | 1.48 | 1.07 | 1.28 | 1.23 | 1.28 | 1.39 | 1.04 |
| histone exchange | 7 | 1.15 | 1.19 | 1.23 | 1.17 | 1.15 | 1.2 | 1.05 |
| ATPase activity, coupled | 283 | -1.01 | 1.06 | 1.16 | 1.03 | 1.13 | 1.15 | 1.04 |
| chaperone binding | 16 | -1.03 | -1.17 | 1.06 | 1.16 | 1.34 | 1.16 | -1.27 |
| response to unfolded protein | 2 | 1.04 | 1.38 | 1.16 | -1.1 | 1.05 | 1.03 | 1.56 |
| protein refolding | 7 | 1.25 | 1.05 | 1.14 | -1.0 | 1.11 | -1.26 | 1.35 |
| motor activity | 76 | -1.02 | 1.07 | 1.2 | 1.02 | 1.14 | 1.1 | 1.03 |
| microtubule motor activity | 51 | 1.03 | 1.09 | 1.15 | 1.06 | 1.12 | 1.1 | 1.04 |
| axoneme | 14 | -1.03 | 1.04 | 1.03 | 1.02 | -1.05 | -1.01 | -1.1 |
| muscle contraction | 12 | 1.16 | -1.04 | 1.06 | -1.18 | 1.13 | 1.33 | 1.25 |
| contractile fiber | 33 | 2.45 | -1.47 | -1.08 | -1.25 | 1.2 | 1.97 | -1.14 |
| actin binding | 114 | -1.07 | 1.06 | 1.38 | -1.0 | 1.31 | 1.09 | 1.15 |
| protein binding, bridging | 17 | -1.01 | 1.02 | 1.14 | 1.03 | 1.09 | 1.16 | 1.1 |
| actin filament binding | 8 | -1.01 | -1.14 | 1.31 | -1.17 | 1.11 | 1.2 | 1.33 |
| actin filament bundle assembly | 17 | 1.57 | -1.02 | 1.24 | -1.1 | 1.24 | 1.29 | 1.51 |
| barbed-end actin filament capping | 2 | -1.38 | -1.03 | 2.18 | 1.01 | 1.59 | -1.39 | -1.04 |
| actin monomer binding | 1 | -3.23 | -1.14 | 1.86 | -1.09 | 2.26 | -4.44 | -1.1 |
| actin filament depolymerization | 4 | -1.06 | 1.03 | 1.27 | 1.14 | 1.25 | -1.37 | -1.31 |
| actin cytoskeleton organization | 160 | 1.17 | 1.02 | 1.34 | -1.03 | 1.29 | 1.3 | 1.17 |
| membrane | 2078 | 1.02 | -1.01 | 1.0 | -1.02 | 1.04 | 1.09 | 1.03 |
| regulation of actin filament length | 20 | -1.48 | 1.08 | 1.45 | 1.05 | 1.38 | -1.21 | 1.18 |
| defense response | 178 | 1.35 | -1.27 | -1.19 | -1.18 | -1.1 | 1.18 | 1.09 |
| acute-phase response | 1 | 1.84 | 1.22 | 1.59 | 1.61 | 1.65 | 1.41 | 1.04 |
| lysozyme activity | 14 | 1.98 | -1.64 | -2.07 | -1.66 | -1.61 | -1.16 | 1.06 |
| defense response to bacterium | 84 | 1.33 | -1.21 | -1.16 | -1.18 | -1.05 | 1.15 | 1.15 |
| male-specific defense response to bacterium | 1 | -1.1 | -1.03 | -1.03 | -1.0 | -1.23 | -1.2 | -1.1 |
| defense response to fungus | 28 | 1.68 | -1.27 | -1.31 | -1.29 | -1.25 | 1.08 | 1.24 |
| response to virus | 12 | 2.35 | -1.64 | -1.09 | -1.61 | 1.07 | 1.84 | 1.26 |
| serine-type endopeptidase activity | 263 | -2.24 | -1.42 | -1.67 | -1.14 | -1.46 | -2.32 | -1.65 |
| blood coagulation | 1 | 1.05 | 1.37 | 1.17 | 1.34 | 1.05 | 1.15 | 1.1 |
| protein-glutamine gamma-glutamyltransferase activity | 2 | 5.84 | -1.94 | -1.03 | -1.59 | -1.03 | 2.32 | 1.28 |
| antigen binding | 1 | -1.71 | -1.18 | -1.3 | -1.15 | -1.23 | -1.3 | 1.07 |
| catalytic activity | 3804 | -1.08 | -1.02 | -1.07 | 1.01 | 1.04 | -1.07 | -1.05 |
| alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity | 1 | 1.32 | -4.96 | -6.02 | -4.98 | -1.62 | 7.64 | -3.94 |
| alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity | 1 | 3.18 | 1.02 | 1.47 | 1.11 | 1.44 | 4.09 | 2.36 |
| beta-1,4-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity | 1 | 3.39 | -1.12 | 1.06 | -1.11 | 1.06 | 1.27 | -1.12 |
| beta-alanyl-dopamine hydrolase activity | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| beta-alanyl-dopamine synthase activity | 1 | 1.36 | 1.09 | 1.02 | -1.03 | -1.16 | 1.04 | 1.01 |
| beta-carotene 15,15'-monooxygenase activity | 1 | 1.12 | -1.01 | 1.14 | 1.06 | 1.09 | 1.14 | -1.06 |
| beta-galactoside alpha-2,6-sialyltransferase activity | 1 | 1.19 | 1.12 | 1.07 | 1.08 | 1.03 | 1.22 | -1.05 |
| beta-ureidopropionase activity | 1 | 17.28 | -3.07 | -1.83 | -2.92 | -2.4 | 3.32 | 1.6 |
| gamma-glutamylcyclotransferase activity | 2 | 1.29 | 1.27 | 1.3 | -1.14 | -1.14 | 1.15 | -1.11 |
| gamma-glutamyltransferase activity | 4 | 1.24 | -1.34 | -1.15 | 1.08 | 1.94 | 1.31 | -1.06 |
| 1-acylglycerol-3-phosphate O-acyltransferase activity | 6 | -1.56 | -1.19 | -1.0 | 1.18 | 2.24 | -2.62 | -1.99 |
| 1-pyrroline-5-carboxylate dehydrogenase activity | 3 | 1.56 | -1.48 | -1.15 | -1.62 | -1.43 | 2.14 | 1.15 |
| 1,3-beta-D-glucan synthase activity | 1 | 12.72 | -1.88 | -1.16 | -3.3 | -1.54 | 3.13 | 2.87 |
| 1,4-alpha-glucan branching enzyme activity | 1 | 3.55 | -1.35 | -1.97 | 1.08 | 1.09 | 1.58 | 1.19 |
| 1-alkyl-2-acetylglycerophosphocholine esterase activity | 1 | 3.85 | 1.07 | 1.8 | 1.09 | 1.57 | 2.16 | 1.8 |
| 2-hydroxyacylsphingosine 1-beta-galactosyltransferase activity | 5 | -4.34 | 1.01 | 1.27 | 1.24 | 1.5 | -3.41 | -1.26 |
| 3-beta-hydroxy-delta5-steroid dehydrogenase activity | 1 | 7.52 | -1.63 | -1.52 | -1.44 | -1.48 | 2.76 | 1.78 |
| 3-hydroxyacyl-CoA dehydrogenase activity | 4 | 1.05 | 1.0 | -1.41 | 1.24 | -1.28 | 1.08 | 1.21 |
| 3-hydroxyisobutyryl-CoA hydrolase activity | 1 | 1.35 | 1.24 | 1.18 | -1.16 | -1.11 | 6.95 | -1.22 |
| 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity | 2 | 1.67 | 1.29 | 1.69 | -1.02 | 1.34 | 1.99 | 1.11 |
| 3-oxo-5-alpha-steroid 4-dehydrogenase activity | 1 | -1.31 | 1.28 | -1.32 | 1.3 | 1.08 | -2.32 | -1.18 |
| 4-aminobutyrate transaminase activity | 1 | -1.88 | 3.07 | -1.92 | -1.52 | -1.3 | -2.05 | 1.06 |
| 4-hydroxyphenylpyruvate dioxygenase activity | 1 | 1.86 | -1.07 | 1.21 | 1.03 | 1.14 | 1.09 | -1.06 |
| 4-nitrophenylphosphatase activity | 9 | -1.32 | 1.18 | -1.3 | 1.06 | 1.01 | -1.31 | 1.07 |
| 5-aminolevulinate synthase activity | 1 | 1.58 | 1.36 | -1.31 | -1.09 | -1.26 | 1.25 | 1.23 |
| 6-phosphofructokinase activity | 1 | 2.63 | -1.22 | -1.22 | 1.02 | -1.12 | 5.11 | 1.3 |
| 6-phosphofructo-2-kinase activity | 1 | 1.09 | -1.34 | 2.59 | -1.04 | 1.14 | 3.38 | -1.06 |
| 6-pyruvoyltetrahydropterin synthase activity | 1 | 2.12 | 1.31 | 1.32 | 1.23 | 1.73 | 1.01 | 2.0 |
| AMP deaminase activity | 1 | 1.21 | 1.11 | 1.3 | -1.14 | 1.24 | 3.4 | 1.06 |
| ATP citrate synthase activity | 4 | -1.02 | -1.02 | -1.04 | -1.09 | -1.21 | 1.05 | -1.32 |
| protein C-terminal carboxyl O-methyltransferase activity | 1 | -2.13 | 1.18 | 1.09 | 1.41 | 1.8 | -1.71 | -1.76 |
| CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity | 1 | 2.1 | -1.04 | 1.71 | -1.1 | 1.9 | -1.29 | 1.13 |
| CDP-diacylglycerol-serine O-phosphatidyltransferase activity | 1 | 1.61 | 1.37 | 2.22 | 1.0 | 1.13 | 1.67 | 1.83 |
| D-amino-acid oxidase activity | 2 | -2.82 | -1.69 | -2.22 | 1.7 | 1.95 | -1.63 | -1.83 |
| DNA (cytosine-5-)-methyltransferase activity | 2 | 1.2 | 1.07 | 1.01 | 1.32 | 1.13 | -1.12 | -1.02 |
| DNA-directed DNA polymerase activity | 18 | 1.05 | 1.09 | 1.17 | 1.07 | 1.16 | 1.1 | -1.1 |
| DNA primase activity | 2 | 1.15 | 1.13 | 1.17 | 1.19 | 1.37 | 1.14 | -1.16 |
| DNA-directed RNA polymerase activity | 28 | 1.31 | 1.12 | 1.19 | 1.24 | 1.24 | 1.06 | 1.0 |
| deoxyribodipyrimidine photo-lyase activity | 1 | -1.34 | -1.01 | -1.03 | 1.23 | 1.09 | -1.47 | -1.1 |
| DNA-(apurinic or apyrimidinic site) lyase activity | 5 | 1.72 | -1.0 | 1.07 | 1.11 | 1.19 | 1.32 | 1.0 |
| methylated-DNA-[protein]-cysteine S-methyltransferase activity | 1 | 1.12 | -1.01 | 1.19 | 1.74 | 1.2 | 1.06 | -1.09 |
| DNA ligase activity | 3 | 1.18 | 1.28 | 1.26 | 1.14 | 1.32 | 1.13 | 1.13 |
| DNA ligase (ATP) activity | 2 | 1.29 | 1.46 | 1.49 | 1.24 | 1.5 | 1.27 | 1.28 |
| DNA photolyase activity | 3 | 1.02 | -1.07 | -1.02 | 1.07 | 1.04 | 1.25 | -1.04 |
| DNA (6-4) photolyase activity | 1 | 1.03 | -1.11 | 1.01 | -1.14 | 1.01 | 1.07 | -1.17 |
| DNA topoisomerase activity | 7 | 1.03 | 1.16 | 1.23 | 1.17 | 1.3 | 1.13 | 1.25 |
| DNA topoisomerase type I activity | 3 | -1.14 | 1.27 | 1.28 | 1.28 | 1.24 | 1.04 | 1.14 |
| DNA topoisomerase (ATP-hydrolyzing) activity | 3 | -1.26 | 1.43 | 1.56 | 1.29 | 1.52 | 1.12 | 1.27 |
| FMN adenylyltransferase activity | 2 | 1.48 | 1.18 | 1.47 | 1.03 | -1.01 | 1.23 | 1.07 |
| GMP synthase activity | 1 | 1.14 | 1.09 | 1.34 | -1.05 | 1.24 | 1.35 | 2.12 |
| GMP synthase (glutamine-hydrolyzing) activity | 1 | 1.14 | 1.09 | 1.34 | -1.05 | 1.24 | 1.35 | 2.12 |
| GPI-anchor transamidase activity | 1 | 1.21 | 1.1 | 1.8 | 1.25 | 1.66 | -1.16 | 1.35 |
| synaptic vesicle uncoating | 2 | -1.44 | 1.22 | 1.71 | 1.06 | 1.91 | 1.53 | 1.42 |
| vesicle-mediated transport | 402 | 1.05 | 1.08 | 1.23 | 1.09 | 1.24 | 1.19 | 1.14 |
| protein import into nucleus | 36 | 1.11 | 1.22 | 1.35 | 1.2 | 1.37 | 1.25 | 1.17 |
| cytoskeleton organization | 516 | 1.14 | 1.08 | 1.28 | 1.06 | 1.28 | 1.2 | 1.11 |
| secretion | 145 | -1.04 | 1.05 | 1.22 | 1.11 | 1.24 | 1.07 | 1.07 |
| GTP cyclohydrolase activity | 1 | 33.5 | -5.63 | -5.79 | -6.03 | -4.58 | -1.32 | 6.94 |
| GTP cyclohydrolase I activity | 1 | 33.5 | -5.63 | -5.79 | -6.03 | -4.58 | -1.32 | 6.94 |
| hydrogen ion transporting ATP synthase activity, rotational mechanism | 16 | 1.35 | -1.04 | 1.11 | -1.09 | -1.01 | 1.54 | -1.13 |
| IMP cyclohydrolase activity | 1 | -1.39 | 1.0 | -1.49 | -1.01 | -1.06 | 1.63 | -1.09 |
| IMP dehydrogenase activity | 1 | 1.16 | 1.34 | 1.12 | 1.03 | 1.08 | -1.1 | 1.99 |
| L-iditol 2-dehydrogenase activity | 3 | 1.17 | -1.39 | -1.37 | 1.12 | 1.23 | 1.74 | -1.02 |
| L-iduronidase activity | 1 | -1.71 | 1.0 | 1.11 | -1.04 | 1.62 | -1.12 | -1.27 |
| N-acetylgalactosamine-4-sulfatase activity | 4 | 1.55 | -1.16 | -1.4 | 1.09 | 1.42 | 2.61 | -1.17 |
| N4-(beta-N-acetylglucosaminyl)-L-asparaginase activity | 3 | -1.38 | 1.01 | 1.13 | 1.06 | 1.15 | -1.4 | 1.08 |
| NAD+ ADP-ribosyltransferase activity | 1 | -2.18 | 1.38 | 1.41 | 1.1 | 1.13 | -1.47 | 1.05 |
| NAD+ kinase activity | 3 | 1.2 | 1.17 | -1.08 | 1.1 | 1.06 | -1.04 | 1.02 |
| NAD+ synthase (glutamine-hydrolyzing) activity | 1 | 2.07 | 1.6 | 1.04 | 1.19 | 1.4 | 2.13 | 1.77 |
| NADH dehydrogenase activity | 36 | 1.13 | 1.03 | 1.27 | 1.06 | 1.1 | 1.72 | -1.29 |
| NAD(P)+-protein-arginine ADP-ribosyltransferase activity | 2 | 1.3 | 1.3 | 1.14 | 1.02 | 1.15 | -1.14 | 1.12 |
| NADPH-hemoprotein reductase activity | 1 | 1.63 | 1.35 | 1.12 | -1.0 | 1.2 | -1.04 | 1.34 |
| NADPH dehydrogenase activity | 1 | -1.51 | 1.45 | 1.08 | 1.06 | -1.12 | -1.58 | -1.42 |
| RNA-3'-phosphate cyclase activity | 2 | 1.22 | 1.13 | -1.15 | 1.27 | 1.07 | -1.19 | -1.17 |
| RNA-directed DNA polymerase activity | 1 | 1.54 | 1.82 | 1.92 | -1.04 | 1.87 | 1.15 | 2.39 |
| UDP-N-acetylglucosamine diphosphorylase activity | 1 | -1.57 | -1.13 | 1.65 | -1.09 | 1.26 | -1.83 | 1.72 |
| UDP-glucose 4-epimerase activity | 3 | -1.33 | 1.08 | -1.17 | -1.19 | -1.1 | -1.47 | 1.1 |
| UDP-glucose:glycoprotein glucosyltransferase activity | 1 | 1.21 | 1.16 | 1.35 | -1.04 | 1.59 | 1.69 | 2.54 |
| UTP:glucose-1-phosphate uridylyltransferase activity | 1 | 1.07 | -1.0 | -1.14 | -1.11 | -1.05 | -1.01 | 1.08 |
| acetyl-CoA C-acetyltransferase activity | 2 | 1.25 | 1.16 | -1.11 | 1.2 | -1.13 | 1.26 | -1.35 |
| acetate-CoA ligase activity | 2 | 1.63 | 1.11 | -1.36 | -1.04 | -1.71 | 1.16 | 1.18 |
| acetyl-CoA C-acyltransferase activity | 4 | 1.58 | 1.11 | -1.25 | 1.12 | 1.08 | 1.06 | -1.32 |
| acetyl-CoA carboxylase activity | 1 | 7.73 | 1.2 | -1.41 | -2.71 | 1.2 | 3.14 | 4.69 |
| acetylcholinesterase activity | 1 | 2.27 | 1.23 | -5.17 | -1.59 | -2.2 | -9.9 | 9.42 |
| acid phosphatase activity | 12 | 1.44 | 1.13 | -1.05 | -1.03 | -1.02 | 1.81 | 1.28 |
| aconitate hydratase activity | 4 | 1.11 | 1.25 | 1.22 | -1.14 | -1.03 | 1.75 | 1.32 |
| acyl-CoA dehydrogenase activity | 15 | -1.17 | 1.11 | -1.37 | 1.1 | 1.1 | -1.79 | -1.3 |
| acyl-CoA oxidase activity | 6 | -2.19 | 1.03 | -1.7 | 1.08 | 1.14 | -3.66 | -1.47 |
| acylphosphatase activity | 6 | 1.57 | 1.0 | -1.02 | 1.01 | -1.1 | 1.04 | -1.12 |
| adenine phosphoribosyltransferase activity | 1 | 1.17 | 1.37 | 1.23 | 1.24 | 1.3 | 1.33 | -1.5 |
| adenosine deaminase activity | 8 | -1.69 | 1.1 | 1.06 | -1.03 | -1.09 | 1.15 | -1.16 |
| adenosine kinase activity | 4 | 1.16 | 1.08 | -1.11 | 1.18 | 1.12 | 1.14 | -1.1 |
| ATP-dependent DNA helicase activity | 19 | 1.12 | 1.17 | 1.33 | 1.14 | 1.3 | 1.15 | 1.17 |
| ATP-dependent RNA helicase activity | 38 | 1.22 | 1.21 | 1.3 | 1.2 | 1.31 | 1.37 | 1.16 |
| cation-transporting ATPase activity | 26 | 1.1 | 1.0 | 1.24 | -1.08 | 1.02 | 1.51 | -1.05 |
| metal ion transmembrane transporter activity | 144 | -1.31 | -1.08 | -1.41 | -1.18 | -1.18 | 1.02 | -1.13 |
| copper-exporting ATPase activity | 1 | -2.22 | 1.33 | 3.02 | 1.07 | -1.16 | -1.12 | 1.08 |
| ATPase activity, coupled to transmembrane movement of substances | 120 | -1.08 | -1.04 | 1.03 | -1.07 | 1.02 | 1.16 | -1.05 |
| ATP-binding cassette (ABC) transporter complex | 41 | 1.23 | -1.2 | -1.24 | -1.08 | 1.0 | 1.04 | -1.04 |
| phospholipid-translocating ATPase activity | 6 | -2.65 | 1.11 | -1.12 | 1.22 | 1.24 | -1.33 | -1.09 |
| adenosylhomocysteinase activity | 3 | -2.93 | -1.82 | -1.21 | -1.07 | 1.22 | 1.86 | -1.16 |
| adenosylmethionine decarboxylase activity | 1 | 1.82 | 1.07 | 1.08 | 1.22 | 1.03 | 1.75 | -1.05 |
| adenylate cyclase activity | 12 | 1.43 | 1.1 | 1.07 | -1.04 | 1.13 | 1.5 | 1.22 |
| adenylate kinase activity | 6 | 1.61 | -1.18 | 1.01 | 1.04 | 1.05 | 1.04 | -1.33 |
| N6-(1,2-dicarboxyethyl)AMP AMP-lyase (fumarate-forming) activity | 1 | 1.1 | 1.08 | 1.31 | 1.11 | 1.27 | 1.3 | 1.19 |
| adenylosuccinate synthase activity | 1 | 2.08 | 1.19 | 1.56 | -1.1 | -1.15 | 1.56 | -1.05 |
| adenylylsulfate kinase activity | 1 | -2.63 | 1.06 | 1.41 | 1.07 | 1.11 | -1.64 | -1.02 |
| L-alanine:2-oxoglutarate aminotransferase activity | 1 | -2.33 | 1.13 | 1.14 | 1.09 | 1.76 | 1.77 | -1.09 |
| alcohol dehydrogenase (NAD) activity | 5 | 1.66 | -1.09 | -1.8 | -1.39 | -1.21 | 1.37 | 1.27 |
| aldehyde dehydrogenase (NAD) activity | 3 | -1.65 | 1.15 | -1.65 | 1.05 | -1.41 | -1.9 | -1.31 |
| aldehyde dehydrogenase [NAD(P)+] activity | 1 | 1.67 | -1.24 | -1.26 | 1.36 | 2.03 | -1.43 | 1.25 |
| alditol:NADP+ 1-oxidoreductase activity | 6 | -1.18 | 1.24 | -1.3 | -1.16 | -1.25 | -1.07 | 1.15 |
| aldo-keto reductase (NADP) activity | 7 | 1.25 | 1.15 | -1.37 | -1.18 | -1.21 | 1.14 | 1.22 |
| aldose 1-epimerase activity | 3 | -1.46 | 1.17 | -1.47 | -1.07 | 1.13 | -1.58 | -1.03 |
| alkaline phosphatase activity | 13 | -5.32 | -1.81 | -4.2 | -1.19 | -1.19 | -4.42 | -4.05 |
| allantoinase activity | 1 | 2.85 | -1.01 | 1.16 | 1.03 | -1.08 | 1.74 | -1.1 |
| amidase activity | 1 | 1.54 | -1.32 | 2.9 | 1.39 | -1.83 | -6.19 | -2.79 |
| amidophosphoribosyltransferase activity | 2 | 1.11 | 1.02 | -1.15 | -1.13 | -1.43 | 1.89 | -1.09 |
| aminoacyl-tRNA hydrolase activity | 3 | 1.72 | -1.02 | 1.14 | 1.1 | 1.0 | 1.49 | -1.08 |
| aminoacylase activity | 6 | -1.72 | -1.86 | -1.07 | -1.63 | 1.29 | -1.6 | -1.71 |
| aminomethyltransferase activity | 4 | 1.27 | 1.14 | 1.17 | 1.01 | -1.01 | 1.6 | 1.08 |
| nucleoside-diphosphatase activity | 3 | -2.29 | 1.44 | -2.09 | 1.33 | -1.38 | -3.32 | 1.1 |
| nucleoside-triphosphatase activity | 535 | 1.04 | 1.07 | 1.18 | 1.06 | 1.16 | 1.12 | 1.03 |
| arginase activity | 2 | 1.73 | -1.09 | 1.02 | -1.05 | -1.18 | 2.27 | 1.17 |
| arginine kinase activity | 3 | 1.01 | 1.07 | 1.29 | 1.05 | -1.11 | 1.23 | -1.22 |
| argininosuccinate synthase activity | 1 | -31.89 | 1.77 | -20.72 | 1.41 | 1.0 | -12.66 | -3.5 |
| argininosuccinate lyase activity | 1 | -15.39 | 1.51 | -7.96 | 1.39 | 1.15 | -4.77 | -1.11 |
| arginyltransferase activity | 1 | 1.03 | 1.39 | 1.46 | 1.24 | 1.37 | 1.69 | 1.36 |
| aromatic-L-amino-acid decarboxylase activity | 4 | 2.04 | -1.66 | -1.74 | -1.59 | -1.59 | 1.51 | 1.84 |
| aralkylamine N-acetyltransferase activity | 1 | -8.98 | 1.77 | -2.05 | -1.02 | -2.08 | -14.22 | 1.22 |
| arylamine N-acetyltransferase activity | 1 | -8.98 | 1.77 | -2.05 | -1.02 | -2.08 | -14.22 | 1.22 |
| aryl sulfotransferase activity | 1 | 1.25 | -1.11 | -1.17 | -1.08 | -1.01 | 1.11 | 1.01 |
| aryldialkylphosphatase activity | 1 | 1.9 | -1.44 | -1.39 | -2.02 | 1.17 | 21.38 | -1.25 |
| asparagine synthase (glutamine-hydrolyzing) activity | 2 | -1.07 | 1.2 | 1.14 | 1.26 | 1.17 | 1.27 | -1.14 |
| asparaginase activity | 4 | 1.31 | -1.05 | -1.24 | -1.15 | 1.04 | 1.97 | 1.03 |
| aspartate 1-decarboxylase activity | 1 | 1.44 | -1.11 | -1.15 | 1.16 | -1.17 | -1.1 | -1.2 |
| L-aspartate:2-oxoglutarate aminotransferase activity | 2 | 1.83 | 1.18 | -1.32 | -1.0 | 1.19 | 1.57 | 1.14 |
| aspartate carbamoyltransferase activity | 1 | -1.12 | 1.14 | 1.63 | 1.21 | 1.33 | 1.0 | 1.05 |
| biliverdin reductase activity | 1 | -1.51 | 1.45 | 1.08 | 1.06 | -1.12 | -1.58 | -1.42 |
| biotin carboxylase activity | 3 | 2.88 | 1.25 | -1.21 | -1.16 | -1.0 | 2.26 | 1.91 |
| biotin-[acetyl-CoA-carboxylase] ligase activity | 1 | 1.43 | 1.46 | 1.44 | 1.24 | 1.28 | 1.18 | 1.38 |
| biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity | 1 | 1.43 | 1.46 | 1.44 | 1.24 | 1.28 | 1.18 | 1.38 |
| bis(5'-nucleosyl)-tetraphosphatase (asymmetrical) activity | 1 | -1.85 | 1.12 | 1.37 | 1.12 | 1.23 | -1.02 | 1.02 |
| branched-chain-amino-acid transaminase activity | 1 | 5.21 | 1.63 | 1.44 | -1.45 | -2.46 | 1.42 | 2.6 |
| carbamoyl-phosphate synthase (ammonia) activity | 1 | -1.12 | 1.14 | 1.63 | 1.21 | 1.33 | 1.0 | 1.05 |
| carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity | 1 | -1.12 | 1.14 | 1.63 | 1.21 | 1.33 | 1.0 | 1.05 |
| carbonate dehydratase activity | 17 | -1.34 | -1.07 | 1.2 | -1.25 | 1.01 | 1.12 | 1.01 |
| carbonyl reductase (NADPH) activity | 3 | 1.57 | 1.0 | -1.52 | -1.25 | -1.09 | 1.43 | 1.09 |
| carboxylesterase activity | 33 | -1.1 | 1.01 | -1.29 | -1.03 | -1.13 | -1.36 | -1.13 |
| carnitine O-acetyltransferase activity | 3 | -1.35 | 1.07 | -1.21 | 1.11 | 1.17 | -1.19 | -1.2 |
| carnitine O-palmitoyltransferase activity | 1 | 1.55 | 1.41 | -1.09 | 1.13 | 1.34 | -1.06 | 1.05 |
| catalase activity | 3 | 1.88 | -1.66 | -2.02 | -1.59 | -1.55 | 2.15 | -1.21 |
| catechol oxidase activity | 1 | 1.73 | 1.33 | 1.02 | 1.13 | -1.06 | -1.05 | 1.06 |
| chitin deacetylase activity | 2 | 2.14 | -1.13 | 1.05 | -1.06 | -1.29 | -1.18 | -1.25 |
| chitin synthase activity | 2 | -4.74 | 1.25 | 1.37 | -1.3 | 1.38 | 1.09 | 1.15 |
| choline O-acetyltransferase activity | 1 | 1.49 | -1.03 | -1.23 | 1.03 | 1.04 | -1.13 | 1.06 |
| choline kinase activity | 2 | 1.11 | 1.13 | 1.36 | 1.19 | 1.43 | 2.57 | 1.13 |
| cholinesterase activity | 2 | -1.12 | 1.28 | -2.63 | -1.13 | -1.28 | -3.75 | 2.43 |
| choline-phosphate cytidylyltransferase activity | 2 | 1.06 | -1.23 | 1.07 | 1.02 | 1.15 | -1.4 | 1.55 |
| citrate (Si)-synthase activity | 1 | -1.05 | 1.1 | -1.11 | 1.07 | -1.23 | -1.06 | -1.08 |
| coproporphyrinogen oxidase activity | 1 | -1.38 | 1.31 | 1.39 | 1.26 | 1.22 | -1.88 | -1.39 |
| cyclic-nucleotide phosphodiesterase activity | 8 | 1.68 | 1.07 | 1.28 | 1.01 | 1.26 | 2.25 | 1.48 |
| 3',5'-cyclic-nucleotide phosphodiesterase activity | 8 | 1.68 | 1.07 | 1.28 | 1.01 | 1.26 | 2.25 | 1.48 |
| 3',5'-cyclic-AMP phosphodiesterase activity | 4 | 1.06 | 1.26 | 1.31 | 1.05 | 1.3 | 1.79 | 1.4 |
| calmodulin-dependent cyclic-nucleotide phosphodiesterase activity | 1 | -1.09 | -1.0 | 1.07 | 1.3 | -1.04 | -1.15 | -1.07 |
| cystathionine beta-synthase activity | 1 | 1.25 | 1.1 | 1.48 | -1.98 | -7.11 | -1.17 | 2.59 |
| cystathionine gamma-lyase activity | 2 | 1.42 | 1.32 | 1.04 | 1.04 | 1.07 | -1.09 | 1.37 |
| cytidine deaminase activity | 3 | -2.8 | 1.04 | -1.39 | 1.2 | 1.32 | -1.87 | -1.06 |
| cytidylate kinase activity | 1 | -1.35 | 1.03 | 1.12 | 1.16 | 1.1 | -1.2 | -1.23 |
| cytochrome-b5 reductase activity | 1 | -1.1 | 1.03 | -1.07 | 1.37 | 1.05 | -1.32 | 1.01 |
| cytochrome-c oxidase activity | 18 | -1.12 | -1.08 | 1.07 | -1.07 | 1.03 | 1.19 | -1.31 |
| cytosine deaminase activity | 1 | 1.58 | 1.21 | -1.04 | 1.33 | -1.0 | -1.93 | 1.34 |
| dCMP deaminase activity | 1 | 2.54 | -1.17 | -1.05 | 1.35 | 1.96 | 1.77 | -1.1 |
| glycogen debranching enzyme activity | 1 | 1.91 | -1.57 | -1.98 | 1.36 | -1.16 | 2.47 | 1.07 |
| 4-alpha-glucanotransferase activity | 1 | 1.91 | -1.57 | -1.98 | 1.36 | -1.16 | 2.47 | 1.07 |
| amylo-alpha-1,6-glucosidase activity | 1 | 1.91 | -1.57 | -1.98 | 1.36 | -1.16 | 2.47 | 1.07 |
| deoxyribose-phosphate aldolase activity | 1 | -1.52 | 1.52 | -1.38 | 1.37 | -1.06 | -2.8 | 1.21 |
| dephospho-CoA kinase activity | 2 | 1.04 | 1.25 | -1.2 | 1.45 | 1.19 | -1.52 | -1.32 |
| diacylglycerol cholinephosphotransferase activity | 1 | -3.65 | 1.65 | 3.17 | 1.08 | 1.15 | -2.09 | 2.51 |
| diacylglycerol kinase activity | 8 | 1.65 | 1.05 | -1.03 | -1.02 | -1.04 | 1.17 | 1.24 |
| diacylglycerol O-acyltransferase activity | 1 | -1.05 | 1.33 | 3.49 | 1.16 | 1.98 | 2.09 | 1.25 |
| dihydrofolate reductase activity | 1 | 2.25 | -1.1 | -1.03 | 1.28 | 1.09 | 2.14 | -1.24 |
| dihydrolipoamide branched chain acyltransferase activity | 1 | 1.74 | 1.44 | 1.67 | 1.01 | 1.35 | 1.38 | 1.19 |
| dihydrolipoyl dehydrogenase activity | 1 | -1.4 | -1.14 | 2.27 | -1.25 | -1.48 | 1.94 | -1.5 |
| dihydrolipoyllysine-residue succinyltransferase activity | 1 | 1.27 | -1.02 | 1.79 | -1.24 | 1.05 | 3.95 | -1.37 |
| dihydroorotase activity | 1 | -1.12 | 1.14 | 1.63 | 1.21 | 1.33 | 1.0 | 1.05 |
| dihydroorotate dehydrogenase activity | 2 | -1.09 | 1.44 | 1.03 | 1.31 | -1.02 | -1.29 | -1.1 |
| 6,7-dihydropteridine reductase activity | 1 | 2.26 | 1.17 | 1.35 | 1.12 | 1.23 | 1.64 | 1.14 |
| dihydropyrimidinase activity | 1 | -1.0 | 1.2 | 1.11 | 1.02 | 1.13 | 1.01 | 1.16 |
| dihydroorotate oxidase activity | 2 | -1.09 | 1.44 | 1.03 | 1.31 | -1.02 | -1.29 | -1.1 |
| dihydrouracil dehydrogenase (NAD+) activity | 1 | -2.4 | 1.52 | -1.09 | 1.53 | -1.18 | -2.54 | -1.54 |
| dimethylallyltranstransferase activity | 2 | 2.38 | 1.13 | 2.21 | -1.02 | 1.58 | 1.4 | 1.7 |
| diphosphomevalonate decarboxylase activity | 1 | 2.36 | -1.19 | -1.1 | 1.02 | -1.17 | 1.09 | 1.24 |
| diphthine synthase activity | 1 | 1.75 | 1.22 | 1.1 | 1.3 | 1.08 | -1.18 | -1.03 |
| dodecenoyl-CoA delta-isomerase activity | 5 | 1.65 | 1.02 | -1.16 | 1.25 | 1.08 | 1.17 | -1.36 |
| dopachrome isomerase activity | 2 | 2.9 | -1.22 | -1.25 | -1.08 | -1.28 | 3.34 | -1.14 |
| dolichol kinase activity | 1 | -2.66 | 1.27 | -1.76 | 1.39 | 1.67 | -4.72 | -1.84 |
| dolichyl-phosphate-mannose-protein mannosyltransferase activity | 2 | 1.57 | 1.01 | 1.07 | -1.18 | -1.11 | 1.61 | 1.23 |
| dUTP diphosphatase activity | 1 | -1.36 | -1.27 | 1.07 | 1.7 | 1.82 | -3.47 | -2.13 |
| peptidyl-lysine modification to hypusine | 2 | 1.15 | 1.07 | 1.04 | 1.07 | 1.11 | 1.08 | -1.05 |
| deoxyhypusine biosynthetic process from spermidine, using deoxyhypusine synthase | 1 | -1.3 | 1.12 | 1.08 | 1.3 | 1.03 | -1.14 | -1.27 |
| ecdysteroid UDP-glucosyltransferase activity | 2 | -2.06 | 1.41 | -1.15 | 1.08 | 1.07 | -2.31 | -1.09 |
| electron-transferring-flavoprotein dehydrogenase activity | 1 | 2.09 | 1.59 | -1.41 | 1.19 | 1.36 | 1.02 | -1.19 |
| endopeptidase activity | 433 | -1.75 | -1.28 | -1.35 | -1.08 | -1.23 | -1.79 | -1.45 |
| ATP-dependent peptidase activity | 3 | -1.2 | 1.29 | 1.23 | 1.11 | 1.07 | 1.26 | 1.04 |
| aminopeptidase activity | 36 | -2.28 | -1.12 | -1.52 | 1.05 | 1.14 | -1.43 | -1.98 |
| metalloexopeptidase activity | 41 | -2.37 | -1.56 | -1.46 | 1.02 | -1.31 | -2.4 | -1.85 |
| carboxypeptidase activity | 35 | -2.91 | -1.85 | -1.55 | -1.01 | -1.63 | -3.14 | -1.86 |
| metallocarboxypeptidase activity | 24 | -4.15 | -2.21 | -1.86 | -1.01 | -1.57 | -4.07 | -2.61 |
| serine-type carboxypeptidase activity | 11 | -1.26 | -1.29 | -1.05 | -1.03 | -1.8 | -1.77 | 1.1 |
| aspartic-type endopeptidase activity | 19 | -1.6 | -1.03 | 1.15 | -1.16 | -1.46 | -1.86 | -1.32 |
| calcium-dependent cysteine-type endopeptidase activity | 4 | 2.12 | 1.16 | 1.44 | 1.14 | 1.21 | 1.24 | 1.16 |
| apoptotic process | 69 | 1.05 | 1.06 | 1.22 | 1.08 | 1.24 | -1.05 | 1.11 |
| lysosome | 28 | -2.58 | -1.1 | -2.35 | 1.15 | -1.08 | -2.06 | -3.03 |
| cysteine-type peptidase activity | 43 | -1.18 | -1.07 | 1.14 | -1.01 | 1.29 | -1.03 | -1.06 |
| dipeptidyl-peptidase activity | 13 | -1.05 | 1.04 | -1.41 | 1.07 | 1.08 | -1.08 | 1.15 |
| pyroglutamyl-peptidase activity | 2 | 2.37 | -1.16 | 1.04 | -1.04 | 1.14 | 1.57 | 1.04 |
| ubiquitin thiolesterase activity | 27 | 1.19 | 1.22 | 1.57 | 1.17 | 1.42 | 1.39 | 1.24 |
| metalloendopeptidase activity | 79 | -1.34 | -1.11 | -1.09 | 1.01 | 1.06 | -1.1 | -1.21 |
| dipeptidase activity | 7 | -1.14 | -1.08 | -1.07 | 1.19 | 1.09 | -1.2 | -1.2 |
| mitochondrion | 480 | 1.05 | 1.07 | 1.13 | 1.06 | 1.11 | 1.38 | -1.19 |
| metallopeptidase activity | 153 | -1.78 | -1.24 | -1.3 | 1.02 | -1.0 | -1.44 | -1.51 |
| peptidyl-dipeptidase activity | 6 | -1.43 | -1.17 | -1.51 | -1.07 | 1.03 | 1.0 | -1.19 |
| vacuole | 54 | -2.1 | -1.04 | -1.32 | -1.01 | -1.13 | -1.28 | -1.9 |
| serine-type peptidase activity | 288 | -2.24 | -1.4 | -1.61 | -1.12 | -1.45 | -2.28 | -1.59 |
| tripeptidyl-peptidase activity | 2 | -2.48 | 1.24 | -1.3 | 1.26 | 1.28 | -1.53 | -1.15 |
| threonine-type endopeptidase activity | 23 | 1.34 | -1.02 | 1.1 | 1.11 | 1.12 | -1.15 | -1.1 |
| peptidase activity | 576 | -1.77 | -1.28 | -1.36 | -1.05 | -1.15 | -1.74 | -1.45 |
| enoyl-CoA hydratase activity | 5 | 1.01 | 1.21 | -1.01 | 1.14 | 1.18 | -1.41 | -1.2 |
| epoxide hydrolase activity | 3 | -2.11 | 1.3 | -1.14 | -1.06 | -1.44 | -4.35 | 1.38 |
| estradiol 17-beta-dehydrogenase activity | 2 | 1.13 | -1.01 | -1.43 | 1.15 | 1.17 | -1.17 | -1.46 |
| ethanolamine kinase activity | 2 | 1.11 | 1.13 | 1.36 | 1.19 | 1.43 | 2.57 | 1.13 |
| ethanolamine-phosphate cytidylyltransferase activity | 1 | 1.81 | -1.85 | 1.07 | -1.42 | 1.03 | 1.7 | 2.82 |
| ethanolaminephosphotransferase activity | 1 | -1.35 | 1.28 | 1.66 | 1.08 | 1.2 | 1.32 | 1.18 |
| farnesyltranstransferase activity | 2 | 1.07 | 1.18 | 1.54 | 1.21 | 1.32 | 1.31 | -1.04 |
| fatty acid synthase activity | 8 | 1.58 | -1.38 | -1.32 | -1.5 | -1.29 | 1.18 | 1.72 |
| [acyl-carrier-protein] S-acetyltransferase activity | 2 | 3.49 | -1.07 | -1.08 | -1.15 | 1.4 | 1.97 | 2.1 |
| [acyl-carrier-protein] S-malonyltransferase activity | 3 | 2.14 | -1.05 | -1.1 | -1.07 | 1.17 | 1.47 | 1.42 |
| 3-oxoacyl-[acyl-carrier-protein] synthase activity | 3 | 2.25 | -1.02 | -1.08 | -1.11 | 1.15 | 1.9 | 1.44 |
| 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) activity | 2 | 3.49 | -1.07 | -1.08 | -1.15 | 1.4 | 1.97 | 2.1 |
| 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase activity | 2 | 3.49 | -1.07 | -1.08 | -1.15 | 1.4 | 1.97 | 2.1 |
| enoyl-[acyl-carrier-protein] reductase (NADPH, B-specific) activity | 2 | 3.49 | -1.07 | -1.08 | -1.15 | 1.4 | 1.97 | 2.1 |
| oleoyl-[acyl-carrier-protein] hydrolase activity | 2 | 3.49 | -1.07 | -1.08 | -1.15 | 1.4 | 1.97 | 2.1 |
| fatty-acyl-CoA synthase activity | 1 | 2.47 | -1.2 | -1.84 | 1.79 | 2.31 | -2.71 | -1.43 |
| ferroxidase activity | 3 | -1.88 | -2.6 | -2.43 | -2.04 | 1.07 | -1.41 | 2.03 |
| iron ion transmembrane transporter activity | 7 | 1.31 | -1.87 | -2.05 | -1.74 | -1.19 | 1.52 | 1.79 |
| ferredoxin-NADP+ reductase activity | 1 | -1.19 | 1.24 | 1.2 | 1.43 | 1.53 | -1.12 | -1.13 |
| tetrahydrofolylpolyglutamate synthase activity | 2 | -1.63 | 1.22 | -1.26 | 1.27 | -1.21 | -1.7 | -1.32 |
| S-(hydroxymethyl)glutathione dehydrogenase activity | 1 | 1.29 | 1.1 | 1.03 | 1.22 | 1.29 | 1.66 | -1.14 |
| formate-tetrahydrofolate ligase activity | 1 | -2.72 | 1.36 | -1.4 | 1.55 | -1.7 | -1.17 | -2.58 |
| fructose-2,6-bisphosphate 2-phosphatase activity | 1 | 1.09 | -1.34 | 2.59 | -1.04 | 1.14 | 3.38 | -1.06 |
| fructose-bisphosphate aldolase activity | 1 | 1.83 | 1.08 | -1.27 | -1.04 | 1.23 | 2.05 | 1.42 |
| fumarate hydratase activity | 4 | -1.03 | 1.03 | -1.06 | -1.09 | -1.22 | 1.2 | -1.26 |
| fumarylacetoacetase activity | 1 | 2.81 | -1.06 | -1.21 | -1.09 | 1.58 | 2.01 | -1.24 |
| geranyltranstransferase activity | 2 | 2.38 | 1.13 | 2.21 | -1.02 | 1.58 | 1.4 | 1.7 |
| glucosamine-6-phosphate deaminase activity | 1 | -1.22 | 1.14 | 1.64 | 1.24 | 2.02 | -2.96 | -2.72 |
| glucosamine 6-phosphate N-acetyltransferase activity | 1 | 1.02 | 1.09 | -1.02 | 1.16 | 1.08 | -1.06 | 2.5 |
| glucose dehydrogenase activity | 4 | 2.07 | -1.33 | -1.32 | -1.38 | -1.39 | 2.15 | -1.49 |
| glucose-6-phosphate dehydrogenase activity | 2 | 1.97 | -1.14 | -2.0 | -1.18 | -1.53 | 1.18 | 1.65 |
| glucose-6-phosphatase activity | 1 | 1.02 | 1.09 | 1.02 | 1.0 | -1.15 | -1.1 | -1.14 |
| glucose-6-phosphate isomerase activity | 1 | 4.0 | 1.26 | -2.38 | -1.3 | -1.5 | 3.87 | 2.36 |
| glucosylceramidase activity | 3 | -3.7 | -1.06 | -2.39 | 1.57 | -1.6 | -3.65 | -2.05 |
| glutamate 5-kinase activity | 1 | -29.94 | 1.93 | -2.31 | -1.05 | -1.13 | -7.63 | 1.24 |
| glutamate-5-semialdehyde dehydrogenase activity | 1 | -29.94 | 1.93 | -2.31 | -1.05 | -1.13 | -7.63 | 1.24 |
| glutamate decarboxylase activity | 2 | 1.13 | -1.03 | -1.11 | 1.07 | -1.17 | -1.16 | -1.18 |
| glutamate dehydrogenase (NAD+) activity | 1 | -1.32 | -1.9 | 1.3 | -2.14 | 3.35 | 4.87 | -4.48 |
| glutamate dehydrogenase [NAD(P)+] activity | 2 | -1.13 | -1.34 | 1.15 | -1.41 | 1.89 | 2.28 | -2.14 |
| glutamate synthase (NADPH) activity | 1 | 2.14 | 1.19 | 1.24 | 1.03 | -1.73 | 3.68 | 2.51 |
| glutamate-ammonia ligase activity | 2 | 1.91 | -1.99 | 2.01 | -1.44 | 1.52 | -1.73 | -1.25 |
| glutamate-cysteine ligase activity | 2 | -1.84 | 1.49 | -1.21 | 1.04 | -1.06 | 1.03 | 1.54 |
| glutamine-fructose-6-phosphate transaminase (isomerizing) activity | 2 | -1.0 | -1.44 | 1.48 | -1.44 | 1.38 | 1.11 | 2.78 |
| glutaryl-CoA dehydrogenase activity | 1 | -1.51 | -1.6 | -1.42 | 1.8 | -1.74 | 2.0 | -2.55 |
| glutathione-disulfide reductase activity | 1 | -1.86 | 1.42 | -1.95 | 1.12 | 1.15 | -1.81 | 1.13 |
| glutathione synthase activity | 1 | -4.56 | 1.3 | -1.17 | 1.44 | -1.72 | -2.63 | -1.17 |
| glutathione transferase activity | 36 | -1.24 | 1.1 | 1.04 | -1.09 | -1.01 | -1.48 | -1.09 |
| glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity | 3 | 1.41 | -1.11 | -1.14 | -1.08 | -1.06 | 1.76 | 1.01 |
| glycerol-3-phosphate O-acyltransferase activity | 2 | 3.16 | -1.05 | -1.54 | 1.04 | 2.3 | -1.34 | 1.05 |
| glycerol-3-phosphate dehydrogenase [NAD+] activity | 2 | 1.2 | 1.12 | -1.49 | 1.16 | 1.79 | -1.21 | -1.08 |
| glycerol-3-phosphate dehydrogenase activity | 5 | -1.02 | -1.05 | 1.01 | 1.08 | 1.13 | 1.19 | -1.09 |
| glycerol kinase activity | 5 | 1.42 | -1.01 | 1.08 | 1.36 | 1.35 | 1.01 | -1.3 |
| glycine hydroxymethyltransferase activity | 1 | 4.17 | -1.15 | 1.23 | 1.12 | 1.08 | 1.16 | -1.19 |
| glycogen (starch) synthase activity | 1 | 3.34 | -1.14 | 1.48 | -1.05 | 1.39 | 2.94 | 1.3 |
| glycine decarboxylation via glycine cleavage system | 1 | 1.73 | -1.01 | 1.7 | -1.04 | 1.38 | 3.31 | -1.03 |
| glycine dehydrogenase (decarboxylating) activity | 1 | -1.37 | -1.08 | -1.16 | -1.21 | -1.14 | 1.68 | -1.06 |
| glycolipid mannosyltransferase activity | 1 | -1.85 | 1.22 | -1.04 | 1.31 | 1.1 | -2.17 | 1.06 |
| glycylpeptide N-tetradecanoyltransferase activity | 1 | -1.49 | 1.29 | 1.71 | 1.24 | 1.7 | 1.05 | 1.0 |
| guanosine-diphosphatase activity | 1 | -1.51 | 1.28 | -1.3 | 1.09 | -1.1 | -3.57 | 1.37 |
| guanylate cyclase activity | 13 | -1.19 | -1.02 | -1.04 | 1.02 | -1.07 | -1.19 | -1.11 |
| guanylate kinase activity | 5 | 3.22 | 1.04 | 1.21 | -1.15 | 1.05 | 2.62 | 1.29 |
| receptor binding | 174 | 1.06 | -1.15 | -1.03 | -1.09 | 1.07 | 1.13 | 1.05 |
| cell adhesion | 161 | 1.22 | -1.06 | 1.07 | -1.06 | 1.21 | 1.38 | 1.25 |
| integral to membrane | 1123 | -1.03 | -1.03 | -1.04 | -1.04 | -1.01 | 1.02 | -1.0 |
| cell adhesion molecule binding | 17 | -1.29 | -1.01 | 1.2 | -1.06 | 1.34 | -1.06 | 1.2 |
| helicase activity | 105 | 1.14 | 1.19 | 1.33 | 1.17 | 1.34 | 1.27 | 1.17 |
| heme oxygenase (decyclizing) activity | 1 | 2.0 | 1.05 | 2.09 | 1.06 | 1.46 | 2.34 | 1.12 |
| heparan sulfate 2-O-sulfotransferase activity | 2 | 2.25 | 1.13 | 1.26 | 1.01 | 1.22 | 1.54 | 1.62 |
| hexaprenyldihydroxybenzoate methyltransferase activity | 1 | -1.1 | 1.09 | 1.22 | 1.29 | 1.3 | 1.24 | -1.16 |
| hexokinase activity | 4 | -1.31 | 1.0 | -1.96 | 1.11 | 1.37 | 1.21 | -1.77 |
| histidine decarboxylase activity | 1 | 1.05 | 1.13 | 1.16 | 1.21 | 1.16 | 1.14 | 1.09 |
| histone acetyltransferase activity | 13 | 1.18 | 1.17 | 1.35 | 1.18 | 1.29 | 1.36 | 1.21 |
| histone deacetylase activity | 8 | 1.29 | 1.14 | 1.07 | 1.21 | 1.24 | 1.41 | 1.17 |
| holocytochrome-c synthase activity | 1 | 1.66 | 1.03 | 1.71 | 1.06 | 1.0 | 2.28 | -1.41 |
| homogentisate 1,2-dioxygenase activity | 1 | -1.11 | -1.1 | -1.19 | 1.12 | -1.15 | -1.2 | -1.15 |
| hyalurononglucosaminidase activity | 1 | 1.16 | 1.5 | 1.47 | 1.26 | 1.67 | 1.16 | 1.31 |
| hydroxyacylglutathione hydrolase activity | 1 | -2.43 | 1.25 | -1.81 | 1.44 | 1.13 | -2.07 | -1.36 |
| hydroxymethylbilane synthase activity | 1 | -1.1 | 1.02 | -1.19 | -1.23 | -1.41 | 1.01 | -1.14 |
| hydroxymethylglutaryl-CoA lyase activity | 1 | -1.23 | 1.82 | 1.48 | 1.33 | 1.52 | 1.63 | 1.08 |
| hydroxymethylglutaryl-CoA synthase activity | 1 | 1.84 | -1.29 | 1.3 | -1.14 | 1.76 | 1.59 | 1.19 |
| iduronate-2-sulfatase activity | 1 | 1.62 | 1.05 | 1.28 | 1.01 | 1.11 | 1.28 | 1.12 |
| inorganic diphosphatase activity | 3 | 1.21 | -1.0 | 1.05 | 1.15 | 1.08 | 1.12 | -1.14 |
| phosphatidylinositol phosphate kinase activity | 6 | -1.3 | 1.22 | 1.48 | 1.12 | 1.58 | 1.08 | 1.01 |
| phosphatidylinositol kinase activity | 5 | -1.09 | 1.17 | 1.62 | 1.11 | 1.75 | 1.32 | 1.12 |
| phosphatidylinositol bisphosphate kinase activity | 3 | -1.46 | 1.15 | 1.53 | 1.18 | 1.77 | -1.09 | -1.14 |
| 1-phosphatidylinositol 4-kinase activity | 2 | 1.44 | 1.21 | 1.76 | 1.02 | 1.71 | 2.28 | 1.63 |
| 1-phosphatidylinositol-4-phosphate 5-kinase activity | 3 | -1.16 | 1.29 | 1.43 | 1.06 | 1.4 | 1.27 | 1.16 |
| phosphatidylinositol phospholipase C activity | 2 | 2.58 | 1.12 | 1.89 | -1.08 | 1.02 | 1.84 | 1.76 |
| phosphatidylinositol bisphosphate phosphatase activity | 3 | 1.28 | 1.18 | 1.39 | 1.05 | 1.29 | 1.32 | 1.32 |
| phosphatidylinositol trisphosphate phosphatase activity | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| phosphatidylinositol monophosphate phosphatase activity | 2 | -1.31 | 1.16 | 1.29 | 1.21 | -1.01 | 1.04 | -1.19 |
| inositol phosphate phosphatase activity | 13 | 1.85 | -1.04 | -1.08 | -1.1 | 1.15 | 1.89 | 1.3 |
| phosphatidylinositol-3-phosphatase activity | 1 | -1.51 | 1.05 | 1.08 | 1.24 | -1.11 | -1.13 | -1.35 |
| inositol-1,4-bisphosphate 1-phosphatase activity | 1 | 3.45 | -1.15 | 1.32 | 1.0 | -1.07 | 1.6 | -1.16 |
| phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity | 1 | -1.07 | 1.22 | 1.09 | 1.37 | -1.04 | -1.17 | -1.02 |
| inositol trisphosphate phosphatase activity | 6 | 1.66 | 1.07 | 1.37 | -1.03 | 1.33 | 2.5 | 1.26 |
| inositol-polyphosphate 5-phosphatase activity | 3 | 1.23 | 1.22 | 1.55 | 1.08 | 1.34 | 1.47 | 1.38 |
| isocitrate dehydrogenase activity | 7 | 1.09 | 1.14 | 1.28 | -1.23 | 1.01 | 1.93 | 1.09 |
| isocitrate dehydrogenase (NAD+) activity | 6 | 1.03 | 1.08 | 1.46 | -1.19 | 1.02 | 2.18 | 1.06 |
| isocitrate dehydrogenase (NADP+) activity | 1 | 1.5 | 1.55 | -1.65 | -1.56 | -1.01 | -1.09 | 1.26 |
| isopentenyl-diphosphate delta-isomerase activity | 1 | 3.32 | -1.02 | 1.27 | 1.26 | 1.27 | 1.92 | 1.53 |
| juvenile-hormone esterase activity | 1 | 1.2 | -1.02 | -1.01 | -1.01 | 1.01 | -1.01 | -1.05 |
| ketohexokinase activity | 4 | 1.16 | 1.19 | 1.03 | 1.01 | 1.17 | 1.1 | -1.01 |
| lactate dehydrogenase activity | 2 | 1.05 | 1.2 | 1.7 | -1.34 | -1.08 | -2.44 | 1.33 |
| L-lactate dehydrogenase activity | 2 | 1.05 | 1.2 | 1.7 | -1.34 | -1.08 | -2.44 | 1.33 |
| lactoylglutathione lyase activity | 1 | -1.59 | 1.09 | -1.01 | 1.22 | -1.05 | -1.39 | -1.43 |
| leukotriene-A4 hydrolase activity | 1 | -1.74 | 1.11 | -2.01 | 1.51 | 1.27 | -1.87 | -1.55 |
| lipoprotein lipase activity | 5 | -2.69 | -1.1 | -1.42 | 1.06 | 1.23 | -1.76 | -1.32 |
| long-chain-acyl-CoA dehydrogenase activity | 2 | 1.45 | 1.45 | -1.3 | -1.02 | 1.11 | 1.33 | -1.15 |
| long-chain fatty acid-CoA ligase activity | 7 | 1.81 | -1.3 | -1.35 | 1.09 | 1.2 | -1.91 | -1.26 |
| lysine N-acetyltransferase activity | 1 | 1.11 | 1.34 | 1.12 | 1.23 | 1.07 | 1.27 | 1.32 |
| malic enzyme activity | 4 | -1.07 | 1.26 | 1.14 | -1.52 | -1.06 | 1.05 | 1.65 |
| malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) activity | 4 | -1.07 | 1.26 | 1.14 | -1.52 | -1.06 | 1.05 | 1.65 |
| mannose-1-phosphate guanylyltransferase activity | 2 | -2.09 | 1.14 | 1.2 | 1.25 | 1.24 | -1.89 | 1.17 |
| mannose-6-phosphate isomerase activity | 2 | 3.41 | 1.03 | 1.33 | -1.05 | -1.02 | 3.9 | 1.9 |
| methenyltetrahydrofolate cyclohydrolase activity | 2 | 1.63 | -1.3 | -1.28 | -1.04 | -1.47 | 2.82 | -1.53 |
| methionine adenosyltransferase activity | 1 | 1.6 | 1.05 | 1.23 | -1.23 | 1.41 | 1.78 | 1.98 |
| methionyl-tRNA formyltransferase activity | 1 | 1.38 | 1.09 | 1.78 | 1.2 | 1.53 | 2.7 | -1.25 |
| mRNA (guanine-N7-)-methyltransferase activity | 1 | 1.22 | 1.12 | 1.34 | 1.19 | 1.11 | -1.08 | 1.0 |
| mRNA guanylyltransferase activity | 1 | 1.69 | 1.28 | 1.3 | 1.3 | 1.17 | 1.5 | 1.34 |
| methylcrotonoyl-CoA carboxylase activity | 2 | 1.38 | 1.49 | 1.32 | 1.28 | 1.37 | 1.61 | 1.17 |
| methylenetetrahydrofolate dehydrogenase [NAD(P)+] activity | 1 | 2.55 | -3.49 | -3.06 | -4.0 | -1.9 | 6.63 | -1.86 |
| methylenetetrahydrofolate dehydrogenase (NAD+) activity | 1 | 7.21 | -2.31 | -1.17 | -1.68 | -1.27 | 9.3 | 1.1 |
| methylenetetrahydrofolate dehydrogenase (NADP+) activity | 2 | 1.63 | -1.3 | -1.28 | -1.04 | -1.47 | 2.82 | -1.53 |
| methylenetetrahydrofolate reductase (NADPH) activity | 1 | 1.73 | -1.17 | -1.14 | -1.13 | -1.11 | 2.12 | -1.29 |
| methylmalonate-semialdehyde dehydrogenase (acylating) activity | 1 | -1.74 | 1.44 | -1.08 | -1.1 | -1.07 | 1.5 | 1.28 |
| mevalonate kinase activity | 1 | 1.42 | 1.16 | 1.14 | 1.22 | 1.05 | 1.2 | -1.02 |
| monooxygenase activity | 35 | 1.52 | -1.2 | -1.23 | -1.13 | -1.16 | -1.04 | 1.06 |
| N,N-dimethylaniline monooxygenase activity | 2 | 1.33 | 1.51 | -1.79 | -1.37 | -1.35 | 1.43 | 1.59 |
| dopamine beta-monooxygenase activity | 3 | 1.25 | 1.07 | -1.02 | -1.01 | 1.04 | 1.12 | 1.05 |
| ecdysone 20-monooxygenase activity | 1 | -1.22 | -1.23 | 1.13 | -1.06 | 1.31 | -1.16 | -1.34 |
| kynurenine 3-monooxygenase activity | 1 | 1.05 | 1.0 | -1.01 | 1.01 | -1.05 | -1.09 | -1.05 |
| monophenol monooxygenase activity | 3 | 1.29 | 1.11 | -1.09 | -1.0 | -1.13 | -1.11 | -1.01 |
| peptidylglycine monooxygenase activity | 1 | 2.74 | 1.3 | 1.75 | 1.08 | 1.98 | 1.97 | 2.28 |
| phenylalanine 4-monooxygenase activity | 2 | 1.24 | -1.3 | -1.46 | -1.42 | -1.18 | 2.11 | -1.36 |
| tryptophan 5-monooxygenase activity | 2 | 1.24 | -1.3 | -1.46 | -1.42 | -1.18 | 2.11 | -1.36 |
| tyrosine 3-monooxygenase activity | 1 | -1.05 | -1.1 | 1.05 | 1.12 | -1.1 | -1.1 | 1.78 |
| inositol-3-phosphate synthase activity | 1 | 1.86 | -1.46 | -1.12 | -1.51 | 1.13 | 1.83 | 1.71 |
| nicotinate-nucleotide diphosphorylase (carboxylating) activity | 1 | 8.31 | 1.05 | -1.79 | -1.54 | -1.19 | 1.17 | 2.54 |
| nicotinate phosphoribosyltransferase activity | 1 | 8.31 | 1.05 | -1.79 | -1.54 | -1.19 | 1.17 | 2.54 |
| nitric-oxide synthase activity | 1 | 1.08 | 1.06 | 1.02 | 1.1 | 1.05 | -1.07 | -1.02 |
| nuclease activity | 79 | 1.35 | 1.08 | 1.23 | 1.13 | 1.28 | 1.26 | 1.07 |
| endonuclease activity | 38 | 1.44 | 1.02 | 1.21 | 1.09 | 1.25 | 1.3 | 1.05 |
| endodeoxyribonuclease activity | 15 | 1.39 | -1.02 | 1.1 | -1.02 | 1.11 | 1.13 | 1.04 |
| endoribonuclease activity | 19 | 1.25 | 1.08 | 1.3 | 1.22 | 1.37 | 1.23 | -1.0 |
| ribonuclease H activity | 2 | -1.13 | 1.13 | 1.07 | 1.39 | 1.3 | -1.08 | -1.14 |
| ribonuclease III activity | 4 | 1.01 | 1.12 | 1.7 | -1.0 | 1.33 | 1.5 | 1.2 |
| ribonuclease P activity | 3 | 1.41 | -1.04 | -1.03 | 1.27 | 1.1 | 1.0 | -1.33 |
| exonuclease activity | 33 | 1.16 | 1.17 | 1.21 | 1.21 | 1.29 | 1.11 | 1.04 |
| exodeoxyribonuclease activity | 7 | -1.09 | 1.11 | 1.04 | 1.05 | 1.14 | -1.05 | -1.01 |
| deoxyribonuclease II activity | 1 | 3.57 | -3.74 | -3.12 | -4.14 | -2.27 | -1.33 | 4.32 |
| exoribonuclease activity | 18 | 1.38 | 1.17 | 1.2 | 1.26 | 1.28 | 1.2 | 1.08 |
| 5'-3' exoribonuclease activity | 2 | -1.11 | 1.53 | 1.16 | 1.33 | 1.34 | -1.11 | 1.19 |
| poly(A)-specific ribonuclease activity | 2 | 1.33 | 1.15 | 1.69 | 1.2 | 1.62 | 2.01 | 1.48 |
| deoxyribonuclease activity | 24 | 1.28 | 1.02 | 1.14 | 1.01 | 1.16 | 1.13 | 1.07 |
| ribonuclease activity | 41 | 1.37 | 1.11 | 1.23 | 1.22 | 1.32 | 1.27 | 1.04 |
| tRNA-specific ribonuclease activity | 4 | 1.4 | 1.02 | -1.04 | 1.26 | 1.1 | -1.04 | -1.24 |
| nucleoside diphosphate kinase activity | 4 | 1.0 | 1.09 | 1.36 | 1.22 | 1.36 | 1.06 | -1.14 |
| nucleotide diphosphatase activity | 2 | 1.11 | 1.19 | 1.38 | 1.17 | 1.36 | 1.15 | -1.0 |
| octanol dehydrogenase activity | 1 | 1.29 | 1.1 | 1.03 | 1.22 | 1.29 | 1.66 | -1.14 |
| hydrolase activity, hydrolyzing O-glycosyl compounds | 93 | -1.83 | -1.29 | -2.4 | -1.22 | -1.47 | -1.78 | -1.26 |
| alpha,alpha-trehalase activity | 3 | -2.73 | 1.23 | -2.68 | 1.17 | -1.22 | -2.41 | 1.01 |
| alpha-amylase activity | 2 | -34.84 | -2.22 | -3.89 | 2.13 | -10.58 | -52.06 | -2.46 |
| alpha-galactosidase activity | 1 | -4.17 | 1.11 | -1.19 | -1.11 | -1.3 | -2.49 | 3.87 |
| alpha-glucosidase activity | 13 | -7.82 | -1.11 | -3.92 | 1.01 | -2.19 | -4.92 | -2.34 |
| alpha-mannosidase activity | 9 | -14.64 | -1.14 | -7.39 | 1.54 | -1.15 | -14.33 | -25.61 |
| alpha-L-fucosidase activity | 1 | -1.1 | -1.19 | -1.53 | -1.9 | -1.86 | -1.28 | 7.99 |
| alpha-N-acetylglucosaminidase activity | 1 | -1.03 | -1.29 | -2.28 | -2.21 | -1.93 | 20.24 | 3.19 |
| beta-N-acetylhexosaminidase activity | 4 | 2.6 | -1.04 | -1.38 | 1.02 | 1.14 | -1.03 | 1.23 |
| beta-galactosidase activity | 2 | -4.22 | 1.94 | -2.65 | -1.31 | -2.65 | -1.75 | -2.68 |
| beta-glucuronidase activity | 2 | -2.98 | 1.22 | 1.8 | 1.28 | 1.95 | -1.65 | 1.15 |
| beta-mannosidase activity | 1 | -5.95 | 1.18 | 1.25 | 1.37 | 1.4 | -3.3 | -1.02 |
| chitinase activity | 17 | -1.5 | -2.2 | -3.34 | -2.35 | -1.62 | 1.11 | 2.06 |
| mannosyl-oligosaccharide 1,2-alpha-mannosidase activity | 5 | 1.56 | 1.24 | -1.27 | -1.11 | -1.07 | 2.0 | 1.29 |
| mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity | 2 | 2.21 | -1.18 | -1.12 | 1.09 | -1.0 | 4.22 | 1.53 |
| mannosyl-oligosaccharide glucosidase activity | 1 | 1.11 | 1.54 | -1.06 | 1.25 | 1.09 | -1.47 | 1.06 |
| oligo-1,6-glucosidase activity | 1 | -7.77 | -4.97 | -3.61 | 4.46 | -5.62 | -8.7 | -6.61 |
| oligosaccharyl transferase activity | 5 | -1.28 | 1.21 | 1.16 | 1.14 | 1.2 | -1.13 | 1.35 |
| dolichyl-diphosphooligosaccharide-protein glycotransferase activity | 2 | -2.3 | 1.16 | -1.02 | 1.14 | 1.1 | -1.42 | 1.33 |
| dolichyl-phosphate beta-glucosyltransferase activity | 1 | -1.18 | 1.16 | 1.9 | 1.06 | 1.5 | -1.14 | 1.68 |
| dolichyl-phosphate beta-D-mannosyltransferase activity | 1 | 1.01 | 1.25 | 1.35 | 1.09 | 1.77 | -1.32 | 1.33 |
| ornithine decarboxylase activity | 2 | -1.3 | -1.56 | -1.58 | -1.19 | -1.51 | 1.68 | -1.8 |
| ornithine-oxo-acid transaminase activity | 2 | -3.39 | 1.33 | -4.62 | 1.16 | 1.17 | -3.49 | 1.2 |
| orotate phosphoribosyltransferase activity | 1 | 1.61 | 1.06 | 1.09 | 1.15 | 1.38 | -1.09 | 1.01 |
| orotidine-5'-phosphate decarboxylase activity | 1 | 1.61 | 1.06 | 1.09 | 1.15 | 1.38 | -1.09 | 1.01 |
| oxoglutarate dehydrogenase (succinyl-transferring) activity | 2 | -2.58 | 1.25 | 1.08 | -1.04 | 1.34 | -1.08 | -1.45 |
| pantothenate kinase activity | 1 | 2.17 | 1.37 | -1.41 | 1.23 | 1.4 | -1.72 | 1.12 |
| pantetheine-phosphate adenylyltransferase activity | 1 | -1.07 | 1.17 | -1.47 | 1.46 | 1.3 | -2.5 | -1.32 |
| peptide alpha-N-acetyltransferase activity | 2 | -1.32 | 1.41 | 1.74 | 1.19 | 1.16 | -1.05 | -1.05 |
| peptide-aspartate beta-dioxygenase activity | 1 | 1.22 | -1.0 | 1.11 | -1.0 | 1.04 | 1.19 | -1.11 |
| cyclosporin A binding | 1 | -1.56 | 1.12 | -1.6 | -1.09 | -1.49 | -1.56 | -1.26 |
| peroxidase activity | 22 | -1.07 | -1.02 | -1.12 | -1.17 | -1.19 | -1.23 | 1.01 |
| glutathione peroxidase activity | 8 | -1.36 | 1.09 | -1.0 | -1.35 | -1.25 | -2.14 | 1.1 |
| phosphatidate cytidylyltransferase activity | 1 | 3.07 | 1.1 | 2.07 | -1.42 | 3.13 | -1.11 | 1.4 |
| phosphatidylserine decarboxylase activity | 1 | 2.68 | -1.15 | 1.36 | 1.05 | 1.26 | 2.6 | 1.26 |
| phosphoacetylglucosamine mutase activity | 1 | -2.86 | 1.51 | 1.12 | 1.09 | 1.09 | -5.18 | 1.1 |
| phosphoenolpyruvate carboxykinase activity | 2 | -2.6 | -1.77 | 1.37 | -1.78 | 2.07 | 1.05 | -1.67 |
| phosphoenolpyruvate carboxykinase (GTP) activity | 2 | -2.6 | -1.77 | 1.37 | -1.78 | 2.07 | 1.05 | -1.67 |
| phosphoglucomutase activity | 1 | 3.54 | -1.2 | -2.31 | -1.28 | -1.28 | 4.52 | 1.34 |
| phosphomannomutase activity | 3 | -1.23 | -1.01 | 1.33 | 1.13 | 1.42 | -1.37 | 1.13 |
| phosphogluconate dehydrogenase (decarboxylating) activity | 4 | 1.7 | 1.24 | -1.62 | -1.03 | -1.05 | 1.36 | 1.59 |
| phosphoglycerate dehydrogenase activity | 2 | 2.82 | -1.08 | -1.11 | 1.42 | -1.05 | 1.77 | -1.02 |
| phosphoglycerate kinase activity | 2 | 1.52 | -1.04 | -1.22 | 1.01 | -1.05 | 1.8 | -1.0 |
| phosphoglycerate mutase activity | 4 | 1.53 | -1.02 | -1.23 | -1.07 | -1.15 | 1.51 | 1.11 |
| phospholipase activity | 42 | -2.02 | -1.32 | -1.4 | -1.24 | -1.44 | -1.94 | -1.19 |
| lysophospholipase activity | 5 | -1.5 | -1.08 | -1.31 | -1.5 | -1.48 | 1.02 | 1.77 |
| phospholipase A2 activity | 10 | 1.72 | -1.05 | -1.16 | -1.1 | -1.05 | -1.02 | 1.07 |
| extracellular region | 527 | -1.12 | -1.32 | -1.37 | -1.26 | -1.26 | -1.16 | 1.03 |
| calcium-dependent phospholipase A2 activity | 1 | -1.04 | -1.03 | -1.01 | -1.13 | -1.15 | 1.09 | -1.0 |
| cytosol | 213 | 1.26 | 1.07 | 1.15 | 1.06 | 1.18 | 1.2 | 1.04 |
| calcium-independent phospholipase A2 activity | 2 | 1.71 | 1.33 | 1.36 | 1.12 | 1.41 | 1.4 | 1.33 |
| phospholipase C activity | 8 | 1.4 | 1.12 | 1.15 | -1.01 | -1.08 | 1.08 | 1.32 |
| phosphomevalonate kinase activity | 1 | 2.16 | -1.1 | 1.03 | -1.13 | 1.06 | 2.2 | 1.18 |
| phosphopantothenate--cysteine ligase activity | 1 | -1.95 | 1.39 | 1.68 | 1.01 | 1.49 | -1.24 | 1.08 |
| phosphopantothenoylcysteine decarboxylase activity | 1 | 3.72 | -1.03 | 1.39 | 1.15 | 1.52 | 2.47 | 1.16 |
| phosphopyruvate hydratase activity | 1 | 1.97 | 1.06 | -1.39 | -1.17 | -1.32 | 2.59 | 1.17 |
| phosphoribosylamine-glycine ligase activity | 1 | 1.35 | 1.17 | 1.22 | 1.29 | 1.23 | 1.38 | 1.07 |
| phosphoribosylaminoimidazole carboxylase activity | 2 | -1.17 | -1.46 | -1.46 | -1.15 | -1.17 | 1.65 | -1.07 |
| phosphoribosylaminoimidazolesuccinocarboxamide synthase activity | 2 | -1.17 | -1.46 | -1.46 | -1.15 | -1.17 | 1.65 | -1.07 |
| phosphoribosylformylglycinamidine cyclo-ligase activity | 1 | 1.35 | 1.17 | 1.22 | 1.29 | 1.23 | 1.38 | 1.07 |
| phosphoribosylformylglycinamidine synthase activity | 1 | -1.91 | 1.54 | -1.81 | 1.0 | -1.5 | -1.22 | 1.08 |
| phosphoribosylaminoimidazolecarboxamide formyltransferase activity | 1 | -1.39 | 1.0 | -1.49 | -1.01 | -1.06 | 1.63 | -1.09 |
| phosphoribosylglycinamide formyltransferase activity | 1 | 1.35 | 1.17 | 1.22 | 1.29 | 1.23 | 1.38 | 1.07 |
| phosphorylase activity | 1 | 2.98 | -1.08 | 1.5 | -1.31 | 1.12 | 4.86 | 1.33 |
| phosphoserine phosphatase activity | 1 | 2.48 | 1.99 | -2.53 | -1.16 | 1.91 | 2.86 | 1.87 |
| O-phospho-L-serine:2-oxoglutarate aminotransferase activity | 1 | 2.63 | -1.02 | -1.9 | 1.11 | -1.04 | 2.51 | 1.93 |
| poly(ADP-ribose) glycohydrolase activity | 1 | 1.16 | 1.03 | 1.19 | 1.34 | 1.67 | -1.1 | 1.01 |
| polynucleotide 5'-phosphatase activity | 1 | 1.69 | 1.28 | 1.3 | 1.3 | 1.17 | 1.5 | 1.34 |
| polynucleotide adenylyltransferase activity | 6 | 1.11 | 1.12 | 1.2 | 1.08 | 1.07 | 1.17 | 1.11 |
| polypeptide N-acetylgalactosaminyltransferase activity | 14 | -1.29 | -1.23 | -1.25 | -1.28 | -1.26 | -1.61 | 1.75 |
| polyribonucleotide nucleotidyltransferase activity | 1 | -1.12 | 1.21 | 1.17 | 1.13 | 1.3 | 1.14 | -1.02 |
| porphobilinogen synthase activity | 1 | -2.33 | 1.65 | -1.72 | 1.07 | -2.09 | -1.28 | -1.14 |
| iron ion transport | 7 | 1.59 | -1.3 | -1.22 | -1.23 | -1.14 | 1.5 | 1.12 |
| procollagen-proline 4-dioxygenase activity | 26 | -1.01 | -1.0 | -1.08 | -1.01 | -1.13 | -1.04 | -1.05 |
| proline dehydrogenase activity | 1 | 2.07 | -7.1 | -4.47 | -7.74 | -3.15 | 31.25 | -1.33 |
| propionyl-CoA carboxylase activity | 1 | 1.48 | 1.28 | 1.32 | 1.05 | 1.07 | 1.16 | -1.01 |
| prenyltransferase activity | 12 | -1.12 | 1.14 | 1.32 | 1.1 | 1.21 | -1.16 | 1.28 |
| protein farnesyltransferase activity | 1 | -1.83 | -1.28 | -1.22 | 1.05 | -1.01 | -1.04 | 2.75 |
| protein geranylgeranyltransferase activity | 6 | -1.57 | 1.19 | 1.22 | 1.14 | 1.21 | -1.62 | 1.21 |
| CAAX-protein geranylgeranyltransferase activity | 2 | -2.28 | 1.15 | 1.11 | 1.06 | 1.05 | -2.22 | 1.78 |
| Rab geranylgeranyltransferase activity | 4 | -1.3 | 1.2 | 1.28 | 1.18 | 1.3 | -1.38 | -1.0 |
| protein C-terminal S-isoprenylcysteine carboxyl O-methyltransferase activity | 1 | -2.13 | 1.18 | 1.09 | 1.41 | 1.8 | -1.71 | -1.76 |
| protein kinase activity | 255 | 1.18 | 1.11 | 1.27 | 1.06 | 1.27 | 1.34 | 1.17 |
| protein histidine kinase activity | 3 | 1.12 | -1.13 | 1.03 | -1.08 | 1.69 | 1.52 | -1.14 |
| protein serine/threonine kinase activity | 183 | 1.12 | 1.13 | 1.33 | 1.08 | 1.3 | 1.3 | 1.15 |
| transmembrane receptor protein serine/threonine kinase activity | 5 | -1.03 | 1.52 | 1.48 | 1.18 | 1.61 | 1.07 | 1.49 |
| DNA-dependent protein kinase activity | 1 | 1.33 | 1.54 | 1.93 | 1.1 | 1.72 | 1.13 | 1.74 |
| AMP-activated protein kinase activity | 3 | 1.06 | 1.32 | 1.11 | 1.03 | -1.06 | 1.31 | 1.11 |
| calmodulin-dependent protein kinase activity | 8 | 2.03 | 1.07 | 1.21 | 1.02 | 1.3 | 2.02 | 1.5 |
| elongation factor-2 kinase activity | 2 | 1.62 | 1.17 | 1.62 | -1.04 | 1.48 | 1.47 | 1.77 |
| myosin light chain kinase activity | 4 | 1.66 | -1.14 | 1.36 | -1.04 | 1.61 | 1.38 | 1.02 |
| phosphorylase kinase activity | 1 | 1.92 | 1.23 | 1.07 | 1.1 | 1.35 | 3.19 | 1.58 |
| cyclic nucleotide-dependent protein kinase activity | 6 | -1.04 | -1.05 | 1.23 | -1.15 | 1.2 | 2.46 | 1.34 |
| cAMP-dependent protein kinase activity | 3 | 1.72 | -1.04 | 1.4 | -1.32 | 1.33 | 4.14 | 1.26 |
| cGMP-dependent protein kinase activity | 3 | -1.85 | -1.05 | 1.07 | 1.01 | 1.09 | 1.46 | 1.42 |
| cyclin-dependent protein kinase activity | 15 | 1.14 | 1.12 | 1.32 | 1.15 | 1.42 | 1.04 | -1.02 |
| protein kinase C activity | 7 | 1.66 | -1.04 | 1.77 | -1.14 | 1.06 | 2.51 | 1.12 |
| calcium-dependent protein kinase C activity | 3 | 1.12 | 1.04 | 2.13 | -1.07 | 1.01 | 3.21 | -1.04 |
| receptor signaling protein serine/threonine kinase activity | 31 | -1.19 | 1.17 | 1.44 | 1.1 | 1.38 | 1.04 | 1.15 |
| G-protein coupled receptor kinase activity | 3 | 1.16 | 1.4 | 1.53 | 1.09 | 1.29 | 1.52 | 1.29 |
| JUN kinase activity | 2 | -1.09 | 1.03 | 1.52 | 1.15 | 1.41 | -1.08 | 1.01 |
| JUN kinase kinase kinase activity | 2 | 1.23 | 1.27 | 1.56 | 1.2 | 1.54 | 1.23 | 1.3 |
| MAP kinase activity | 9 | -1.46 | 1.12 | 1.25 | 1.14 | 1.35 | -1.25 | 1.02 |
| MAP kinase kinase activity | 3 | -1.23 | 1.23 | 1.34 | 1.19 | 1.4 | 1.25 | 1.23 |
| MAP kinase kinase kinase activity | 5 | -1.39 | 1.21 | 1.48 | 1.15 | 1.82 | 1.09 | 1.04 |
| MAPK/ERK kinase kinase activity | 1 | 1.83 | 1.46 | 1.17 | -1.02 | 1.24 | 1.79 | 1.33 |
| ribosomal protein S6 kinase activity | 1 | -2.62 | 1.78 | 1.7 | 1.33 | 1.66 | 1.52 | 1.3 |
| protein serine/threonine/tyrosine kinase activity | 7 | -1.0 | 1.17 | 1.32 | 1.17 | 1.33 | 1.18 | 1.15 |
| protein tyrosine kinase activity | 43 | 1.39 | 1.05 | 1.14 | 1.01 | 1.19 | 1.42 | 1.33 |
| transmembrane receptor protein tyrosine kinase activity | 21 | 1.55 | -1.07 | 1.14 | -1.01 | 1.13 | 1.58 | 1.42 |
| non-membrane spanning protein tyrosine kinase activity | 7 | 1.37 | 1.16 | 1.77 | 1.12 | 1.46 | 1.64 | 1.42 |
| receptor signaling protein tyrosine kinase activity | 3 | -1.0 | 1.08 | -1.13 | -1.05 | -1.16 | -1.03 | -1.05 |
| protein-L-isoaspartate (D-aspartate) O-methyltransferase activity | 1 | 2.93 | 1.03 | 1.2 | 1.19 | 1.05 | 1.97 | 1.02 |
| protein-lysine 6-oxidase activity | 2 | -1.01 | 1.14 | -1.1 | 1.01 | -1.1 | -1.06 | -1.07 |
| phosphoprotein phosphatase activity | 86 | 1.24 | 1.12 | 1.24 | 1.06 | 1.2 | 1.31 | 1.16 |
| protein serine/threonine phosphatase activity | 42 | 1.25 | 1.09 | 1.29 | 1.04 | 1.19 | 1.37 | 1.15 |
| calcium-dependent protein serine/threonine phosphatase activity | 6 | 1.52 | -1.02 | 1.2 | 1.05 | 1.13 | 1.24 | 1.06 |
| protein tyrosine phosphatase activity | 39 | 1.24 | 1.12 | 1.28 | 1.06 | 1.27 | 1.3 | 1.17 |
| non-membrane spanning protein tyrosine phosphatase activity | 5 | 1.49 | 1.12 | 1.2 | 1.1 | 1.4 | 1.22 | 1.1 |
| prenylated protein tyrosine phosphatase activity | 1 | 1.54 | 1.27 | 2.19 | -1.23 | 1.74 | 3.0 | 1.51 |
| receptor signaling protein tyrosine phosphatase activity | 3 | 2.8 | -1.06 | 1.33 | -1.06 | 1.35 | 3.49 | 1.47 |
| oxygen-dependent protoporphyrinogen oxidase activity | 2 | -1.11 | 1.24 | 1.36 | 1.38 | 1.3 | 1.17 | -1.16 |
| pseudouridylate synthase activity | 3 | 1.57 | 1.31 | 1.17 | 1.55 | 1.45 | 1.03 | 1.08 |
| purine-nucleoside phosphorylase activity | 2 | 1.43 | 1.02 | 1.21 | 1.12 | 1.16 | -1.08 | 1.07 |
| pyridoxamine-phosphate oxidase activity | 3 | 1.93 | 1.06 | -1.17 | 1.32 | 1.07 | 1.07 | -1.07 |
| pyrimidodiazepine synthase activity | 4 | 1.29 | 1.17 | 1.1 | 1.04 | 1.18 | -1.03 | 1.04 |
| pyrroline-5-carboxylate reductase activity | 2 | -1.77 | 1.27 | -1.94 | 1.07 | -1.03 | -1.14 | -1.01 |
| pyruvate carboxylase activity | 1 | 2.41 | -1.07 | -1.67 | 1.11 | -2.11 | 1.65 | 1.08 |
| pyruvate dehydrogenase activity | 3 | -1.02 | 1.01 | 1.31 | 1.1 | -1.0 | 1.57 | -1.25 |
| pyruvate dehydrogenase (acetyl-transferring) activity | 3 | -1.02 | 1.01 | 1.31 | 1.1 | -1.0 | 1.57 | -1.25 |
| pyruvate dehydrogenase (acetyl-transferring) kinase activity | 1 | 1.67 | -1.54 | 1.2 | -1.13 | 7.02 | 3.44 | 1.09 |
| [pyruvate dehydrogenase (lipoamide)] phosphatase activity | 1 | 1.78 | 1.13 | 1.46 | 1.23 | 1.08 | 1.92 | 1.02 |
| dihydrolipoyllysine-residue acetyltransferase activity | 1 | 1.23 | 1.19 | 1.96 | -1.07 | 1.35 | 3.33 | 1.11 |
| pyruvate kinase activity | 7 | 1.18 | 1.01 | 1.0 | -1.0 | -1.04 | 1.2 | -1.0 |
| retinal isomerase activity | 1 | 1.12 | -1.01 | 1.14 | 1.06 | 1.09 | 1.14 | -1.06 |
| retinol dehydrogenase activity | 1 | -1.07 | 1.02 | 1.03 | -1.03 | -1.28 | 1.11 | -1.06 |
| ribokinase activity | 2 | -1.21 | 1.07 | -1.3 | 1.1 | -1.27 | -1.38 | -1.12 |
| ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor | 2 | -2.27 | -1.06 | 1.44 | 1.36 | 1.87 | -1.61 | -1.54 |
| ribose phosphate diphosphokinase activity | 2 | 2.41 | 1.04 | 1.7 | -1.06 | 1.1 | 2.9 | 1.34 |
| ribulose-phosphate 3-epimerase activity | 1 | 3.69 | 1.12 | -1.44 | 1.15 | 1.15 | 1.38 | 1.6 |
| ribose-5-phosphate isomerase activity | 1 | 2.8 | 1.42 | -1.73 | 1.16 | 1.1 | 1.26 | 1.25 |
| saccharopine dehydrogenase activity | 1 | 1.21 | 1.36 | -1.07 | 1.24 | -1.06 | 1.55 | 1.4 |
| saccharopine dehydrogenase (NAD+, L-lysine-forming) activity | 1 | 1.21 | 1.36 | -1.07 | 1.24 | -1.06 | 1.55 | 1.4 |
| selenide, water dikinase activity | 2 | 1.76 | 1.25 | 1.23 | 1.04 | 1.04 | 1.62 | -1.07 |
| sepiapterin reductase activity | 3 | 2.14 | -1.76 | -1.79 | -1.65 | -1.36 | 2.93 | -1.29 |
| serine C-palmitoyltransferase activity | 2 | -2.24 | 1.07 | 2.48 | 1.17 | 1.31 | -1.71 | -1.18 |
| serine-pyruvate transaminase activity | 1 | 1.1 | -1.81 | -2.08 | -1.81 | -1.6 | 3.21 | -1.83 |
| spermidine synthase activity | 2 | 1.05 | 1.2 | 1.25 | 1.18 | 1.33 | 1.44 | -1.0 |
| sphingomyelin phosphodiesterase activity | 5 | -5.07 | -3.51 | -4.56 | 1.49 | -1.12 | -3.62 | -4.08 |
| stearoyl-CoA 9-desaturase activity | 7 | -1.4 | 1.15 | -1.2 | 1.26 | 1.01 | -1.67 | -1.18 |
| sterol esterase activity | 1 | -139.1 | -18.6 | -1.99 | 2.08 | -24.32 | -152.96 | -197.4 |
| sterol O-acyltransferase activity | 3 | -2.3 | -1.64 | -1.24 | 1.46 | 1.26 | -1.59 | -2.16 |
| succinate-CoA ligase activity | 6 | 1.06 | -1.06 | 1.08 | -1.08 | 1.02 | 1.3 | -1.31 |
| succinate-CoA ligase (ADP-forming) activity | 5 | 1.09 | -1.04 | 1.18 | -1.15 | -1.09 | 1.43 | -1.27 |
| succinate-CoA ligase (GDP-forming) activity | 3 | 1.16 | -1.04 | -1.07 | -1.04 | 1.11 | 1.22 | -1.42 |
| succinate-semialdehyde dehydrogenase activity | 1 | -2.19 | 1.37 | 2.06 | 1.34 | 1.0 | 1.58 | -1.11 |
| sulfate adenylyltransferase activity | 1 | -2.63 | 1.06 | 1.41 | 1.07 | 1.11 | -1.64 | -1.02 |
| sulfate adenylyltransferase (ATP) activity | 1 | -2.63 | 1.06 | 1.41 | 1.07 | 1.11 | -1.64 | -1.02 |
| sulfinoalanine decarboxylase activity | 1 | -1.56 | 1.66 | -7.28 | 1.14 | -1.03 | -3.47 | 1.18 |
| superoxide dismutase activity | 5 | 1.45 | 1.15 | -1.15 | 1.12 | -1.07 | 1.37 | -1.06 |
| thiamine diphosphokinase activity | 2 | -1.04 | 1.3 | 1.02 | 1.32 | 1.14 | -1.17 | -1.06 |
| thioredoxin-disulfide reductase activity | 2 | -1.32 | 1.3 | -1.35 | 1.1 | 1.1 | -1.38 | 1.15 |
| thiosulfate sulfurtransferase activity | 1 | 1.15 | -1.38 | 2.62 | 1.38 | 1.04 | -1.42 | -1.83 |
| threonine aldolase activity | 1 | 1.44 | -1.1 | -1.93 | 1.6 | -1.06 | 1.8 | -2.26 |
| L-threonine ammonia-lyase activity | 1 | -3.81 | -2.74 | -2.14 | -1.15 | 11.07 | -1.69 | -3.46 |
| thymidine kinase activity | 1 | 1.02 | 1.09 | 1.32 | 1.32 | 1.3 | -1.29 | -1.13 |
| thymidylate kinase activity | 1 | 1.27 | 1.13 | 1.91 | 1.21 | 1.41 | -1.17 | -1.08 |
| thymidylate synthase activity | 1 | -1.63 | -1.08 | 1.19 | 1.46 | 1.72 | -2.2 | -2.17 |
| sedoheptulose-7-phosphate:D-glyceraldehyde-3-phosphate glyceronetransferase activity | 1 | 3.18 | 1.11 | -3.27 | 1.02 | -1.44 | -1.1 | 1.46 |
| transketolase activity | 2 | 2.03 | 1.17 | -1.56 | 1.02 | 1.12 | 1.44 | 1.62 |
| transposase activity | 1 | 1.07 | -1.04 | -1.16 | 1.02 | 1.48 | -1.27 | -1.22 |
| trehalose-phosphatase activity | 2 | 5.88 | -2.14 | -1.49 | -1.62 | 1.02 | 4.6 | 1.12 |
| triglyceride lipase activity | 38 | -3.83 | -1.33 | -1.88 | -1.09 | -1.77 | -4.41 | -2.22 |
| tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase activity | 1 | 1.03 | 1.15 | 1.06 | 1.36 | -1.06 | -1.46 | -1.19 |
| tRNA (guanine-N2-)-methyltransferase activity | 1 | 1.09 | 1.31 | 1.08 | 1.21 | 1.15 | 1.13 | 1.18 |
| aminoacyl-tRNA ligase activity | 40 | -1.07 | 1.26 | 1.26 | 1.14 | 1.18 | 1.05 | 1.09 |
| alanine-tRNA ligase activity | 3 | -1.05 | 1.37 | 1.07 | 1.17 | 1.09 | 1.18 | 1.07 |
| arginine-tRNA ligase activity | 3 | 1.35 | 1.15 | 1.32 | 1.15 | 1.24 | 1.15 | 1.05 |
| aspartate-tRNA ligase activity | 1 | -1.97 | 1.61 | 1.28 | -1.18 | 1.45 | -1.88 | 1.38 |
| asparagine-tRNA ligase activity | 2 | -1.04 | 1.15 | 1.12 | 1.09 | 1.19 | -1.05 | 1.03 |
| cysteine-tRNA ligase activity | 2 | -1.01 | 1.37 | 1.43 | 1.08 | 1.22 | 1.24 | 1.76 |
| glutamate-tRNA ligase activity | 3 | -1.23 | 1.31 | 1.44 | 1.01 | 1.37 | 1.11 | 1.37 |
| glutamine-tRNA ligase activity | 1 | 1.03 | 1.27 | 1.03 | 1.21 | 1.05 | 1.24 | 1.09 |
| glycine-tRNA ligase activity | 3 | 1.01 | 1.17 | 1.37 | 1.21 | 1.21 | 1.27 | -1.07 |
| histidine-tRNA ligase activity | 1 | -1.32 | 1.37 | -1.07 | 1.31 | -1.14 | -1.74 | -1.19 |
| isoleucine-tRNA ligase activity | 2 | 1.35 | 1.22 | 1.54 | 1.02 | 1.38 | 1.61 | 1.38 |
| leucine-tRNA ligase activity | 2 | -1.17 | 1.4 | 1.1 | 1.12 | 1.07 | 1.1 | 1.24 |
| lysine-tRNA ligase activity | 1 | 1.12 | 1.29 | 1.39 | 1.11 | 1.29 | 1.02 | 1.14 |
| methionine-tRNA ligase activity | 2 | -1.59 | 1.18 | 1.28 | 1.38 | 1.17 | -1.27 | -1.03 |
| phenylalanine-tRNA ligase activity | 3 | -1.16 | 1.27 | 1.38 | 1.24 | 1.13 | -1.0 | -1.14 |
| proline-tRNA ligase activity | 2 | -1.08 | 1.19 | 1.31 | 1.06 | 1.32 | 1.47 | 1.15 |
| serine-tRNA ligase activity | 3 | -1.3 | 1.34 | 1.25 | 1.17 | 1.28 | -1.22 | -1.0 |
| threonine-tRNA ligase activity | 2 | -1.65 | 1.24 | 1.06 | 1.13 | -1.08 | -1.17 | 1.02 |
| tryptophan-tRNA ligase activity | 2 | 1.09 | 1.09 | 1.24 | 1.11 | 1.12 | 1.12 | 1.03 |
| tyrosine-tRNA ligase activity | 2 | 1.14 | 1.27 | 1.29 | 1.14 | 1.19 | 1.03 | -1.04 |
| valine-tRNA ligase activity | 2 | -1.62 | 1.31 | 1.29 | 1.15 | 1.04 | -1.25 | 1.17 |
| tryptophan 2,3-dioxygenase activity | 1 | 1.33 | -1.46 | -1.36 | -1.56 | -1.39 | 2.84 | -1.33 |
| tubulin-tyrosine ligase activity | 11 | 1.17 | 1.01 | -1.01 | 1.04 | -1.02 | 1.1 | 1.04 |
| tyrosine decarboxylase activity | 2 | 2.12 | -1.17 | -1.18 | -1.17 | 1.11 | 1.9 | -1.07 |
| L-tyrosine:2-oxoglutarate aminotransferase activity | 1 | 1.14 | 1.12 | -1.09 | -1.15 | 1.09 | -1.15 | -1.05 |
| ubiquitin activating enzyme activity | 3 | 1.07 | 1.19 | 1.42 | 1.25 | 1.55 | 1.31 | 1.05 |
| ubiquitin-protein ligase activity | 83 | 1.15 | 1.18 | 1.45 | 1.14 | 1.34 | 1.23 | 1.22 |
| ubiquitin-specific protease activity | 8 | 1.17 | 1.21 | 1.8 | 1.18 | 1.58 | 1.96 | 1.41 |
| uracil DNA N-glycosylase activity | 2 | 2.91 | -1.23 | -1.13 | -1.34 | 1.04 | 2.88 | 1.17 |
| uracil phosphoribosyltransferase activity | 1 | 1.85 | 1.12 | 1.41 | 1.03 | 1.01 | 1.67 | 1.33 |
| urate oxidase activity | 1 | 3.87 | -2.08 | -2.11 | -2.0 | -1.26 | -1.05 | -1.64 |
| uridine kinase activity | 2 | 4.18 | -1.72 | 1.72 | -1.65 | -1.27 | 3.76 | 1.43 |
| uridine phosphorylase activity | 3 | -2.2 | -1.73 | 1.35 | -1.47 | -1.37 | -2.19 | 1.39 |
| uroporphyrinogen-III synthase activity | 2 | 1.15 | -1.11 | -1.12 | 1.02 | -1.09 | 1.11 | -1.25 |
| uroporphyrinogen decarboxylase activity | 1 | 2.27 | -1.08 | -1.25 | 1.43 | 1.04 | 1.18 | 1.26 |
| xanthine dehydrogenase activity | 2 | 1.25 | 1.78 | -1.17 | -1.32 | -1.19 | -1.3 | 1.66 |
| xanthine oxidase activity | 1 | 1.38 | 1.39 | -1.09 | 1.59 | 1.39 | -1.89 | 1.34 |
| xylulokinase activity | 2 | 1.23 | 1.04 | -1.16 | -1.08 | 1.26 | 1.41 | 1.22 |
| enzyme inhibitor activity | 101 | 1.28 | -1.39 | -1.39 | -1.37 | -1.17 | 1.33 | 1.03 |
| protein kinase inhibitor activity | 6 | 1.32 | 1.13 | 1.13 | 1.05 | 1.2 | 1.27 | 1.35 |
| cyclin-dependent protein kinase inhibitor activity | 2 | 1.97 | -1.0 | 1.05 | 1.02 | 1.15 | 1.58 | 1.04 |
| protein phosphatase inhibitor activity | 7 | 1.59 | -1.08 | -1.01 | -1.03 | 1.27 | 1.27 | -1.01 |
| protein serine/threonine phosphatase inhibitor activity | 1 | 1.57 | 1.1 | -1.11 | 1.22 | 1.13 | -1.01 | -1.25 |
| endopeptidase inhibitor activity | 73 | 1.41 | -1.48 | -1.45 | -1.49 | -1.3 | 1.51 | 1.04 |
| serine-type endopeptidase inhibitor activity | 58 | 1.2 | -1.48 | -1.57 | -1.54 | -1.37 | 1.38 | -1.05 |
| cysteine-type endopeptidase inhibitor activity | 6 | 3.11 | -1.78 | -1.05 | -1.54 | -1.18 | 1.44 | 1.41 |
| signal transducer activity | 443 | 1.09 | 1.01 | 1.1 | 1.0 | 1.08 | 1.12 | 1.06 |
| receptor activity | 428 | 1.12 | -1.03 | 1.04 | -1.02 | 1.04 | 1.12 | 1.04 |
| ecdysteroid hormone receptor activity | 2 | 1.39 | 1.12 | 1.31 | 1.09 | 1.22 | 1.45 | 1.35 |
| thyroid hormone receptor activity | 2 | -1.28 | 1.12 | 1.08 | -1.12 | -1.1 | -1.03 | -1.04 |
| transmembrane signaling receptor activity | 332 | 1.1 | -1.02 | 1.05 | -1.02 | 1.03 | 1.1 | 1.04 |
| acetylcholine-activated cation-selective channel activity | 9 | -1.07 | 1.01 | -1.02 | -1.03 | -1.09 | -1.06 | -1.03 |
| GABA-A receptor activity | 3 | 1.07 | 1.04 | 1.03 | -1.04 | -1.11 | -1.02 | -1.07 |
| carbohydrate binding | 166 | -1.92 | -1.36 | -1.53 | -1.23 | -1.33 | -1.85 | -1.01 |
| cytokine receptor activity | 1 | 1.28 | 1.03 | 1.08 | -1.21 | -1.11 | 1.23 | -1.01 |
| boss receptor activity | 1 | 2.09 | -1.24 | -1.34 | -1.08 | -1.33 | -1.62 | 1.07 |
| Wnt-activated receptor activity | 5 | 1.17 | 1.03 | 1.52 | -1.14 | 1.66 | -1.04 | 1.31 |
| Wnt receptor signaling pathway | 56 | 1.24 | 1.03 | 1.2 | -1.03 | 1.29 | 1.38 | 1.28 |
| Wnt receptor signaling pathway, calcium modulating pathway | 2 | 1.15 | 1.15 | 1.16 | 1.02 | 1.3 | 1.14 | 2.54 |
| G-protein coupled receptor activity | 168 | 1.06 | -1.03 | 1.05 | -1.02 | 1.01 | 1.02 | -1.02 |
| adrenergic receptor activity | 2 | -1.09 | 1.2 | -1.08 | -1.0 | -1.18 | -1.21 | -1.07 |
| bombesin receptor activity | 2 | 1.03 | 1.12 | 1.37 | -1.03 | -1.15 | 1.42 | 1.13 |
| calcitonin receptor activity | 5 | 1.36 | -1.25 | 1.11 | -1.21 | 1.02 | -1.07 | 1.56 |
| cholecystokinin receptor activity | 2 | -1.2 | -1.05 | 1.02 | -1.05 | -1.23 | -1.25 | -1.17 |
| dopamine receptor activity | 4 | -1.05 | 1.04 | -1.1 | 1.03 | -1.07 | -1.17 | -1.14 |
| G-protein coupled GABA receptor activity | 4 | -1.05 | 1.06 | -1.02 | 1.01 | -1.05 | -1.07 | -1.16 |
| galanin receptor activity | 1 | -1.14 | 1.16 | 1.12 | 1.07 | 1.38 | -1.13 | 1.15 |
| gonadotropin-releasing hormone receptor activity | 2 | 1.14 | 1.03 | 1.06 | 1.02 | -1.01 | -1.0 | -1.03 |
| ionotropic glutamate receptor activity | 35 | 1.11 | -1.01 | 1.02 | -1.04 | -1.06 | -1.02 | -1.03 |
| alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity | 2 | -1.07 | -1.07 | -1.18 | -1.06 | -1.13 | -1.05 | -1.13 |
| N-methyl-D-aspartate selective glutamate receptor activity | 5 | -2.2 | 1.03 | -1.01 | -1.04 | -1.02 | -1.52 | 1.05 |
| leukotriene receptor activity | 1 | 1.34 | 1.16 | 1.19 | 1.06 | 1.04 | 1.4 | -1.06 |
| neuropeptide Y receptor activity | 6 | -1.12 | -1.28 | 1.15 | -1.08 | 1.23 | 2.07 | -1.01 |
| olfactory receptor activity | 58 | -1.05 | 1.02 | -1.02 | 1.02 | -1.07 | -1.04 | -1.08 |
| opioid receptor activity | 1 | 1.11 | 1.13 | -1.05 | 1.1 | 1.01 | 1.15 | 1.06 |
| octopamine receptor activity | 6 | 1.21 | 1.06 | 1.02 | 1.02 | 1.02 | 1.0 | 1.01 |
| serotonin receptor activity | 3 | 1.27 | -1.02 | 1.07 | 1.0 | 1.01 | 1.11 | 1.03 |
| somatostatin receptor activity | 2 | -1.22 | 1.13 | 1.03 | -1.13 | -1.24 | -1.37 | -1.14 |
| tachykinin receptor activity | 6 | -1.02 | -1.12 | 1.07 | -1.2 | -1.05 | 1.79 | 1.44 |
| vasopressin receptor activity | 1 | -1.03 | 1.09 | 1.19 | 1.15 | 1.1 | -1.1 | 1.13 |
| transmembrane receptor protein tyrosine phosphatase activity | 6 | 1.57 | 1.05 | 1.23 | 1.03 | 1.35 | 2.22 | 1.25 |
| ephrin receptor activity | 1 | 2.31 | 1.1 | 1.68 | 1.08 | 2.32 | 2.06 | 2.34 |
| epidermal growth factor-activated receptor activity | 1 | 2.54 | -1.11 | -1.09 | 1.14 | 1.59 | 3.23 | 1.89 |
| fibroblast growth factor-activated receptor activity | 3 | 1.8 | -1.38 | 2.01 | 1.04 | -1.27 | -1.03 | -1.21 |
| insulin-activated receptor activity | 1 | 1.01 | 1.43 | 1.57 | 1.5 | 2.27 | 1.72 | 1.64 |
| insulin-like growth factor-activated receptor activity | 2 | -1.08 | -1.38 | -1.23 | -1.3 | -1.24 | 5.4 | 3.24 |
| signal transduction | 876 | 1.14 | 1.03 | 1.16 | 1.02 | 1.16 | 1.16 | 1.12 |
| growth factor binding | 5 | -1.0 | 1.2 | 1.22 | 1.22 | 1.63 | 1.18 | 1.29 |
| platelet-derived growth factor-activated receptor activity | 1 | -1.05 | 1.27 | 1.32 | -1.04 | 1.15 | 1.36 | 1.38 |
| vascular endothelial growth factor-activated receptor activity | 1 | -1.05 | 1.27 | 1.32 | -1.04 | 1.15 | 1.36 | 1.38 |
| transforming growth factor beta-activated receptor activity | 5 | -1.03 | 1.52 | 1.48 | 1.18 | 1.61 | 1.07 | 1.49 |
| transforming growth factor beta receptor activity, type I | 3 | -1.44 | 1.44 | 1.63 | 1.24 | 1.98 | 1.37 | 1.58 |
| transforming growth factor beta receptor activity, type II | 2 | 1.6 | 1.65 | 1.28 | 1.09 | 1.17 | -1.35 | 1.36 |
| tumor necrosis factor-activated receptor activity | 1 | 10.08 | -2.23 | 1.27 | -2.86 | -1.97 | 3.13 | 1.07 |
| neurotrophin receptor activity | 1 | -1.26 | -1.0 | -1.26 | -1.09 | -1.23 | -1.44 | -1.14 |
| osmosensor activity | 1 | -6.38 | -3.77 | -11.1 | -5.2 | -1.19 | 28.07 | -1.66 |
| death receptor activity | 1 | 10.08 | -2.23 | 1.27 | -2.86 | -1.97 | 3.13 | 1.07 |
| death receptor binding | 1 | 1.78 | 1.11 | 1.56 | 1.24 | 1.76 | -1.03 | 1.11 |
| low-density lipoprotein receptor activity | 9 | 1.21 | 1.15 | 1.29 | -1.02 | -1.05 | 1.37 | 1.08 |
| netrin receptor activity | 2 | -1.2 | 1.24 | 1.26 | 1.34 | 2.11 | -1.46 | 1.04 |
| netrin receptor activity involved in chemorepulsion | 1 | 1.29 | 1.16 | 1.2 | 1.17 | 1.22 | 1.22 | 1.2 |
| scavenger receptor activity | 24 | 1.07 | -1.29 | -1.24 | -1.03 | -1.04 | -1.04 | -1.17 |
| endoplasmic reticulum | 211 | -1.05 | 1.06 | 1.07 | 1.02 | 1.06 | -1.08 | 1.12 |
| KDEL sequence binding | 1 | -1.48 | 1.27 | 1.01 | 1.14 | -1.01 | -1.37 | 1.53 |
| signal recognition particle binding | 3 | -1.47 | 1.36 | 1.04 | 1.08 | -1.23 | -1.47 | 1.4 |
| signal sequence binding | 10 | -1.31 | 1.23 | -1.06 | 1.11 | -1.02 | -1.42 | 1.25 |
| nuclear export signal receptor activity | 3 | 1.18 | 1.05 | 2.95 | 1.07 | 1.92 | 2.19 | 1.34 |
| peroxisome | 39 | 1.3 | -1.06 | -1.53 | 1.05 | 1.16 | -1.5 | -1.16 |
| peroxisome matrix targeting signal-2 binding | 2 | -1.03 | -1.06 | -1.0 | -1.0 | 1.01 | -1.02 | 1.0 |
| peroxisomal membrane | 14 | 1.87 | 1.14 | -1.32 | 1.28 | 1.36 | -1.33 | -1.09 |
| laminin receptor activity | 1 | -9.63 | 1.35 | 1.66 | 1.05 | 1.52 | -7.44 | 1.13 |
| receptor signaling protein activity | 45 | -1.03 | 1.15 | 1.35 | 1.07 | 1.29 | 1.13 | 1.16 |
| transmembrane receptor protein tyrosine kinase signaling pathway | 107 | 1.24 | 1.04 | 1.28 | 1.05 | 1.21 | 1.28 | 1.23 |
| intracellular signal transduction | 268 | 1.18 | 1.1 | 1.26 | 1.06 | 1.23 | 1.24 | 1.17 |
| SH3/SH2 adaptor activity | 9 | 1.2 | -1.01 | 1.23 | 1.07 | 1.17 | 1.44 | -1.06 |
| transmembrane receptor protein serine/threonine kinase signaling pathway | 44 | 1.2 | 1.13 | 1.35 | 1.05 | 1.35 | 1.29 | 1.24 |
| transforming growth factor beta receptor, cytoplasmic mediator activity | 4 | 1.26 | 1.21 | 1.59 | 1.07 | 1.32 | 1.36 | 1.28 |
| transforming growth factor beta receptor, common-partner cytoplasmic mediator activity | 1 | 1.02 | 1.23 | 1.11 | -1.09 | -1.09 | -1.03 | -1.05 |
| transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity | 1 | 2.8 | 1.17 | 2.77 | -1.14 | 1.51 | 2.43 | 1.42 |
| transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity | 1 | -2.15 | 1.12 | -1.08 | 1.29 | -1.07 | -2.29 | 1.33 |
| protein localization | 361 | 1.08 | 1.14 | 1.29 | 1.1 | 1.27 | 1.17 | 1.16 |
| protein kinase A binding | 3 | -1.23 | 1.19 | 1.19 | 1.03 | 1.27 | 1.0 | 1.5 |
| protein kinase C binding | 5 | 1.76 | 1.33 | 1.56 | 1.07 | 1.48 | 1.92 | 1.83 |
| small GTPase regulator activity | 92 | 1.05 | 1.14 | 1.38 | 1.04 | 1.28 | 1.25 | 1.21 |
| guanyl-nucleotide exchange factor activity | 52 | 1.11 | 1.15 | 1.38 | 1.01 | 1.29 | 1.52 | 1.26 |
| ARF guanyl-nucleotide exchange factor activity | 6 | -1.22 | 1.21 | 1.27 | -1.01 | 1.25 | 1.49 | 1.58 |
| Ran guanyl-nucleotide exchange factor activity | 1 | -1.45 | 1.36 | 1.25 | 1.07 | 1.01 | -1.05 | -1.05 |
| Ras guanyl-nucleotide exchange factor activity | 28 | 1.05 | 1.14 | 1.41 | 1.02 | 1.33 | 1.5 | 1.21 |
| Rho guanyl-nucleotide exchange factor activity | 21 | 1.01 | 1.13 | 1.41 | 1.07 | 1.35 | 1.47 | 1.21 |
| GDP-dissociation inhibitor activity | 2 | -1.01 | -1.04 | 1.34 | 1.09 | 1.21 | -1.08 | -1.54 |
| Rab GDP-dissociation inhibitor activity | 1 | 1.43 | 1.0 | 1.11 | 1.08 | 1.12 | 1.1 | -1.39 |
| Rho GDP-dissociation inhibitor activity | 1 | -1.46 | -1.08 | 1.63 | 1.09 | 1.3 | -1.27 | -1.71 |
| GTPase activator activity | 62 | 1.02 | 1.15 | 1.36 | 1.05 | 1.23 | 1.09 | 1.2 |
| Rab GTPase activator activity | 27 | 1.13 | 1.14 | 1.41 | 1.07 | 1.24 | 1.21 | 1.16 |
| Ran GTPase activator activity | 1 | 1.34 | 1.36 | 1.62 | 1.42 | 1.91 | 1.35 | 1.2 |
| Ras GTPase activator activity | 41 | 1.16 | 1.16 | 1.42 | 1.06 | 1.27 | 1.21 | 1.19 |
| Rho GTPase activator activity | 7 | 1.29 | 1.18 | 1.47 | 1.05 | 1.37 | 1.25 | 1.31 |
| fibroblast growth factor receptor binding | 3 | 1.57 | 1.01 | 2.07 | -1.12 | 1.29 | 2.72 | 1.05 |
| ephrin receptor binding | 1 | 1.81 | 1.29 | 1.78 | 1.1 | 1.94 | 2.77 | 1.53 |
| anchored to plasma membrane | 3 | 3.1 | -2.01 | -3.22 | -2.43 | -1.37 | 2.44 | 1.37 |
| integral to plasma membrane | 194 | 1.07 | -1.04 | 1.01 | -1.03 | 1.0 | 1.05 | 1.06 |
| frizzled binding | 3 | -1.65 | -1.07 | 1.18 | -1.29 | 1.31 | 1.28 | 1.51 |
| frizzled-2 binding | 2 | -1.8 | 1.01 | 1.43 | -1.3 | -1.11 | 1.61 | 2.1 |
| Notch binding | 13 | -1.08 | 1.09 | 1.12 | 1.16 | 1.26 | 1.32 | 1.32 |
| patched binding | 2 | -1.41 | -1.16 | 1.11 | 1.42 | 1.34 | -1.32 | -2.31 |
| sevenless binding | 2 | 2.05 | 1.08 | 1.52 | 1.1 | 1.59 | 1.98 | 1.31 |
| smoothened binding | 1 | 1.33 | 1.48 | 1.27 | 1.38 | 1.17 | 1.23 | 1.56 |
| Toll binding | 6 | 2.0 | -1.13 | -1.18 | -1.1 | -1.05 | 3.64 | 1.27 |
| torso binding | 2 | 4.82 | -2.22 | -1.24 | -1.06 | 1.12 | 2.59 | 1.13 |
| cytokine activity | 3 | 2.09 | -1.14 | -1.06 | -1.07 | 1.06 | 1.89 | 1.58 |
| cytokine receptor binding | 13 | 1.33 | 1.01 | -1.03 | -1.2 | 1.15 | 1.04 | 1.28 |
| epidermal growth factor receptor binding | 8 | 2.02 | -1.02 | 1.33 | 1.03 | 1.43 | 2.13 | 1.51 |
| transmembrane receptor protein tyrosine kinase activator activity | 1 | 1.23 | 1.27 | 1.15 | 1.19 | 1.2 | 1.15 | 1.07 |
| positive regulation of epidermal growth factor-activated receptor activity | 1 | -1.21 | 1.16 | 2.16 | -1.11 | 1.4 | 1.68 | 1.37 |
| negative regulation of epidermal growth factor-activated receptor activity | 3 | 1.29 | 1.07 | 1.31 | 1.06 | 1.37 | 1.41 | 1.15 |
| insulin receptor binding | 12 | 1.04 | 1.01 | 1.16 | 1.14 | 1.03 | 1.08 | 1.07 |
| insulin-like growth factor receptor binding | 1 | 2.22 | 1.3 | 2.31 | -1.1 | 1.87 | 2.8 | 1.91 |
| transforming growth factor beta receptor binding | 7 | 1.67 | 1.01 | -1.0 | -1.48 | -1.16 | 1.33 | 1.27 |
| tumor necrosis factor receptor binding | 1 | 1.09 | -1.11 | -1.93 | -1.35 | 4.12 | -1.38 | 6.18 |
| neurotrophin receptor binding | 1 | 1.14 | 1.02 | -1.14 | -1.05 | -1.16 | -1.16 | -1.16 |
| vascular endothelial growth factor receptor binding | 3 | -1.25 | -1.01 | -1.01 | 1.19 | 1.41 | -1.38 | -1.06 |
| neurexin family protein binding | 4 | 1.03 | 1.04 | -1.12 | -1.02 | -1.15 | -1.08 | -1.04 |
| integrin binding | 2 | -1.2 | -1.12 | -1.15 | -1.24 | -1.4 | -1.2 | -1.15 |
| hormone activity | 52 | -1.48 | -1.33 | -1.19 | -1.09 | -1.16 | -1.59 | -1.16 |
| neuropeptide hormone activity | 35 | -1.76 | -1.5 | -1.27 | -1.18 | -1.19 | -1.89 | -1.23 |
| nutrient reservoir activity | 6 | 1.04 | 1.0 | -1.18 | -1.1 | -1.23 | -1.11 | 1.17 |
| structural molecule activity | 451 | 1.11 | -1.08 | -1.04 | -1.04 | -1.01 | 1.08 | -1.06 |
| structural constituent of cell wall | 2 | 4.29 | -1.81 | -1.38 | -1.64 | -1.67 | 1.06 | 1.01 |
| structural constituent of cytoskeleton | 43 | 1.03 | 1.02 | 1.24 | 1.01 | 1.24 | 1.12 | 1.03 |
| extracellular matrix structural constituent | 4 | -1.38 | -1.31 | 1.16 | -1.34 | -1.07 | -1.08 | 1.23 |
| collagen | 3 | 1.14 | -1.43 | 1.08 | -1.25 | -1.21 | 1.82 | -1.14 |
| structural constituent of eye lens | 1 | -23.09 | -8.59 | -4.86 | 2.62 | -1.02 | -36.83 | -31.54 |
| structural constituent of chorion | 9 | 1.18 | 1.05 | -1.08 | 1.04 | -1.21 | -1.1 | -1.03 |
| structural constituent of chitin-based cuticle | 129 | 1.11 | -1.12 | -1.19 | -1.13 | -1.23 | -1.09 | -1.11 |
| transporter activity | 817 | -1.14 | -1.04 | -1.14 | -1.08 | -1.06 | 1.05 | -1.07 |
| ion channel activity | 205 | 1.06 | -1.01 | 1.0 | -1.05 | -1.04 | 1.07 | -1.01 |
| intracellular ligand-gated ion channel activity | 7 | 1.42 | 1.03 | 1.28 | 1.0 | 1.1 | 1.59 | 1.16 |
| intracellular ligand-gated calcium channel activity | 3 | 1.97 | -1.03 | 1.18 | 1.08 | 1.09 | 1.76 | 1.17 |
| ryanodine-sensitive calcium-release channel activity | 1 | 3.82 | -1.03 | 1.3 | -1.12 | 1.16 | 2.73 | 1.18 |
| inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 2.52 | 1.29 | 3.07 | -1.16 | 1.37 | 5.13 | 2.97 |
| intracellular cyclic nucleotide activated cation channel activity | 3 | -1.19 | 1.0 | 1.03 | -1.02 | 1.02 | -1.02 | -1.19 |
| volume-sensitive anion channel activity | 2 | 1.92 | 1.08 | -1.06 | -1.08 | 2.11 | 2.25 | 1.09 |
| calcium activated cation channel activity | 3 | -1.25 | -1.55 | 1.25 | 1.49 | -1.39 | -1.46 | -1.49 |
| extracellular ligand-gated ion channel activity | 58 | 1.01 | 1.01 | -1.01 | -1.09 | -1.14 | -1.1 | 1.02 |
| excitatory extracellular ligand-gated ion channel activity | 51 | 1.09 | 1.0 | -1.01 | -1.09 | -1.16 | -1.07 | 1.03 |
| extracellular-glutamate-gated ion channel activity | 34 | 1.25 | -1.01 | 1.01 | -1.04 | -1.06 | 1.04 | -1.03 |
| inhibitory extracellular ligand-gated ion channel activity | 4 | -1.54 | 1.26 | 1.47 | -1.43 | -1.72 | -1.63 | -1.04 |
| inward rectifier potassium channel activity | 4 | -1.18 | -1.13 | -1.58 | -3.21 | -3.46 | 1.87 | -1.7 |
| gap junction channel activity | 6 | -1.73 | -1.09 | -1.05 | -1.09 | -1.25 | -1.21 | 1.35 |
| voltage-gated ion channel activity | 44 | 1.07 | -1.01 | 1.02 | -1.09 | -1.09 | 1.15 | -1.09 |
| voltage-gated calcium channel activity | 8 | 1.1 | -1.05 | -1.01 | -1.01 | -1.07 | 1.04 | -1.11 |
| calcium channel regulator activity | 2 | -1.03 | -1.06 | -1.05 | -1.01 | -1.02 | -1.08 | -1.24 |
| voltage-gated chloride channel activity | 3 | -1.31 | 1.23 | -1.16 | 1.19 | 1.05 | 1.88 | 1.52 |
| voltage-gated sodium channel activity | 2 | 1.16 | 1.04 | 1.11 | 1.14 | 1.08 | 1.02 | 1.02 |
| voltage-gated potassium channel activity | 25 | 1.14 | -1.03 | 1.01 | -1.2 | -1.15 | 1.11 | -1.15 |
| A-type (transient outward) potassium channel activity | 2 | 3.38 | -1.08 | 1.04 | -1.08 | 1.22 | 1.8 | -1.38 |
| open rectifier potassium channel activity | 3 | 2.34 | -1.1 | -1.13 | -1.16 | 1.03 | 1.04 | -1.38 |
| anion channel activity | 23 | -1.01 | 1.02 | -1.01 | -1.14 | -1.11 | 1.24 | 1.08 |
| chloride channel activity | 13 | -1.14 | 1.15 | 1.04 | -1.13 | -1.12 | 1.12 | 1.12 |
| cation channel activity | 105 | 1.11 | -1.02 | 1.01 | -1.05 | -1.0 | 1.12 | -1.03 |
| calcium channel activity | 24 | 1.39 | -1.04 | 1.09 | -1.06 | 1.15 | 1.41 | 1.14 |
| potassium channel activity | 31 | 1.1 | -1.02 | -1.0 | -1.15 | -1.14 | 1.09 | -1.12 |
| sodium channel activity | 32 | -1.0 | -1.01 | -1.01 | 1.05 | 1.03 | 1.01 | -1.07 |
| amine transmembrane transporter activity | 58 | -1.3 | -1.08 | -1.61 | -1.23 | -1.04 | -1.08 | -1.11 |
| vesicular hydrogen:amino acid antiporter activity | 1 | -1.42 | -1.04 | 1.11 | -1.1 | -1.1 | -1.22 | -1.18 |
| acetylcholine transmembrane transporter activity | 1 | 1.16 | 1.14 | 1.1 | 1.05 | 1.08 | 1.11 | 1.02 |
| acetylcholine:hydrogen antiporter activity | 1 | 1.16 | 1.14 | 1.1 | 1.05 | 1.08 | 1.11 | 1.02 |
| hydrogen:amino acid symporter activity | 2 | 1.74 | 1.43 | -1.72 | -1.09 | 1.17 | -1.0 | 1.38 |
| amino acid transmembrane transporter activity | 53 | -1.39 | -1.07 | -1.67 | -1.24 | -1.03 | -1.11 | -1.1 |
| neutral L-amino acid secondary active transmembrane transporter activity | 1 | -1.01 | 1.0 | 1.27 | 1.1 | -1.12 | 1.05 | -1.06 |
| proline:sodium symporter activity | 1 | 1.01 | -1.09 | -1.18 | -1.18 | 1.11 | -1.08 | -1.12 |
| L-tyrosine transmembrane transporter activity | 2 | 3.0 | -1.1 | 1.15 | -1.28 | -1.42 | 2.49 | 1.13 |
| transmembrane transporter activity | 713 | -1.15 | -1.03 | -1.13 | -1.08 | -1.07 | 1.06 | -1.07 |
| dicarboxylic acid transmembrane transporter activity | 13 | 1.1 | -1.01 | -1.14 | -1.45 | -1.24 | 1.31 | 1.09 |
| organic acid:sodium symporter activity | 8 | -1.17 | 1.19 | -1.85 | -1.42 | -1.96 | -1.64 | 1.15 |
| tricarboxylic acid transmembrane transporter activity | 10 | 1.18 | 1.18 | -1.1 | -1.17 | -1.22 | 1.3 | 1.11 |
| L-glutamate transmembrane transporter activity | 4 | -2.07 | 1.33 | -5.17 | -2.48 | -1.62 | -1.72 | -2.15 |
| high-affinity glutamate transmembrane transporter activity | 1 | -51.3 | 1.56 | -15.93 | -1.24 | 2.78 | -3.27 | -134.18 |
| inorganic phosphate transmembrane transporter activity | 3 | 2.26 | -1.45 | 1.37 | -1.34 | 1.47 | 3.74 | -1.03 |
| high affinity inorganic phosphate:sodium symporter activity | 18 | -3.67 | 1.02 | -3.81 | -1.03 | -1.98 | -1.81 | -2.84 |
| lipid transporter activity | 29 | -1.34 | -1.01 | -1.21 | -1.16 | -1.06 | -1.03 | -1.19 |
| neurotransmitter transporter activity | 24 | 1.11 | -1.3 | -1.53 | -1.28 | 1.11 | 1.77 | 1.29 |
| neurotransmitter:sodium symporter activity | 18 | 1.0 | -1.4 | -1.72 | -1.34 | 1.19 | 1.93 | 1.23 |
| dopamine transmembrane transporter activity | 1 | -1.02 | 1.06 | -1.1 | 1.08 | -1.08 | 1.0 | -1.01 |
| dopamine:sodium symporter activity | 1 | -1.02 | 1.06 | -1.1 | 1.08 | -1.08 | 1.0 | -1.01 |
| gamma-aminobutyric acid:sodium symporter activity | 1 | 1.23 | 1.09 | 1.07 | 1.18 | 1.17 | -1.0 | -1.09 |
| nucleotide-sugar transmembrane transporter activity | 8 | 1.01 | 1.23 | -1.01 | 1.24 | 1.15 | -1.17 | 1.39 |
| organic acid transmembrane transporter activity | 88 | -1.4 | -1.03 | -1.4 | -1.13 | -1.01 | -1.05 | -1.12 |
| sugar:hydrogen symporter activity | 8 | 1.06 | -1.15 | -1.17 | -1.12 | -1.18 | -1.05 | 1.82 |
| fructose transmembrane transporter activity | 3 | 1.33 | -1.15 | -1.45 | -1.42 | -1.39 | -1.07 | -1.47 |
| glucose transmembrane transporter activity | 15 | -1.29 | -1.15 | -1.91 | -1.13 | -1.42 | -1.88 | 1.23 |
| zinc ion transport | 2 | -1.19 | 1.21 | 1.46 | 1.02 | -1.06 | 1.08 | -1.05 |
| taurine transmembrane transporter activity | 1 | 1.14 | 1.06 | -1.1 | -1.01 | -1.05 | 1.05 | 1.32 |
| taurine:sodium symporter activity | 1 | 1.14 | 1.06 | -1.1 | -1.01 | -1.05 | 1.05 | 1.32 |
| tricarboxylate secondary active transmembrane transporter activity | 3 | -1.58 | 1.64 | -1.92 | -1.18 | -1.42 | 1.07 | 1.16 |
| water transmembrane transporter activity | 7 | -2.13 | 1.26 | -1.34 | -1.07 | -2.07 | -2.5 | -1.23 |
| secondary active transmembrane transporter activity | 155 | -1.44 | -1.11 | -1.62 | -1.18 | -1.13 | 1.21 | -1.17 |
| copper ion transmembrane transporter activity | 4 | -1.47 | 1.29 | 1.37 | 1.11 | 1.01 | -1.04 | 1.08 |
| manganese ion transmembrane transporter activity | 2 | -1.53 | 1.28 | -1.11 | 1.02 | -1.23 | -1.54 | 1.12 |
| zinc ion transmembrane transporter activity | 7 | -1.46 | 1.19 | 1.4 | 1.14 | 1.09 | -1.16 | -1.34 |
| calcium-transporting ATPase activity | 4 | -1.18 | -1.01 | 1.34 | -1.06 | 1.25 | -1.0 | 1.03 |
| sodium:potassium-exchanging ATPase activity | 8 | 1.13 | -1.11 | 1.22 | -1.35 | -1.0 | 1.53 | 1.06 |
| eye pigment precursor transporter activity | 3 | 1.41 | -1.1 | -1.26 | -1.07 | -1.12 | -1.06 | -1.05 |
| cation:sugar symporter activity | 8 | 1.06 | -1.15 | -1.17 | -1.12 | -1.18 | -1.05 | 1.82 |
| nucleoside:sodium symporter activity | 2 | -7.59 | 1.47 | -2.97 | -1.0 | -2.3 | -5.85 | -2.0 |
| cation:amino acid symporter activity | 13 | 1.13 | -1.27 | -2.32 | -1.57 | 1.11 | 1.18 | 1.23 |
| proton-dependent oligopeptide secondary active transmembrane transporter activity | 3 | 1.69 | -1.17 | -1.51 | -1.15 | -1.07 | 1.17 | 1.2 |
| synaptic vesicle amine transmembrane transporter activity | 1 | 9.63 | -2.21 | -1.97 | -1.79 | -1.94 | -1.46 | -1.99 |
| calcium:sodium antiporter activity | 2 | -1.37 | 1.7 | 1.3 | -1.23 | 1.65 | 2.01 | 1.96 |
| sodium:phosphate symporter activity | 21 | -3.19 | 1.03 | -3.12 | -1.03 | -1.89 | -1.53 | -2.45 |
| monovalent cation:hydrogen antiporter activity | 4 | -2.37 | -1.01 | -1.35 | 1.01 | -1.74 | 3.33 | -1.27 |
| inorganic anion exchanger activity | 2 | -3.68 | -3.74 | 3.73 | -4.0 | -1.27 | -1.18 | -1.53 |
| CMP-N-acetylneuraminate transmembrane transporter activity | 3 | -1.73 | 1.25 | -1.03 | 1.28 | -1.06 | -1.84 | 1.23 |
| GDP-fucose transmembrane transporter activity | 3 | -1.11 | 1.21 | 1.01 | 1.37 | 1.12 | -1.24 | 1.06 |
| GDP-mannose transmembrane transporter activity | 3 | -1.11 | 1.21 | 1.01 | 1.37 | 1.12 | -1.24 | 1.06 |
| UDP-galactose transmembrane transporter activity | 6 | -1.21 | 1.24 | 1.0 | 1.15 | 1.18 | -1.35 | 1.53 |
| UDP-glucose transmembrane transporter activity | 3 | -1.11 | 1.21 | 1.01 | 1.37 | 1.12 | -1.24 | 1.06 |
| UDP-glucuronic acid transmembrane transporter activity | 4 | -1.46 | 1.25 | 1.0 | 1.26 | 1.04 | -1.57 | 1.43 |
| UDP-N-acetylglucosamine transmembrane transporter activity | 4 | 1.24 | 1.2 | -1.03 | 1.43 | 1.23 | -1.03 | 1.1 |
| UDP-N-acetylgalactosamine transmembrane transporter activity | 2 | -2.31 | 1.25 | -1.08 | 1.21 | 1.09 | -2.5 | 1.48 |
| UDP-xylose transmembrane transporter activity | 3 | -1.4 | 1.3 | 1.05 | 1.18 | -1.06 | -1.45 | 1.9 |
| ATP:ADP antiporter activity | 2 | -1.19 | 1.19 | 1.41 | 1.1 | 1.28 | 1.45 | -1.24 |
| carnitine:acyl carnitine antiporter activity | 2 | 1.34 | 1.32 | -1.91 | 1.22 | 1.14 | -1.7 | -1.23 |
| vacuole organization | 13 | 1.03 | 1.12 | 1.06 | 1.24 | 1.09 | 1.02 | 1.01 |
| vesicle fusion | 8 | -1.38 | -1.08 | 1.22 | 1.35 | 1.26 | -1.28 | -1.21 |
| synaptic vesicle to endosome fusion | 1 | 1.73 | -1.06 | 2.32 | 1.09 | 1.39 | 3.26 | 1.36 |
| vesicle targeting | 12 | -1.08 | 1.24 | 1.4 | 1.15 | 1.31 | 1.23 | 1.25 |
| synaptic vesicle targeting | 9 | 1.07 | 1.25 | 1.5 | 1.16 | 1.39 | 1.39 | 1.23 |
| soluble NSF attachment protein activity | 6 | 1.55 | 1.06 | 1.18 | 1.07 | 1.23 | 1.28 | 1.16 |
| SNAP receptor activity | 20 | 1.03 | 1.15 | 1.31 | 1.2 | 1.21 | -1.11 | 1.1 |
| nucleocytoplasmic transporter activity | 2 | 1.05 | 1.3 | -1.0 | 1.23 | 1.14 | 1.08 | -1.04 |
| binding | 4696 | 1.06 | 1.02 | 1.07 | 1.04 | 1.11 | 1.08 | 1.06 |
| steroid binding | 19 | -2.28 | -1.49 | -1.79 | 1.11 | -1.16 | -2.3 | -2.31 |
| juvenile hormone binding | 2 | -1.14 | 1.17 | 1.21 | 1.18 | 1.14 | 1.31 | 1.23 |
| retinoid binding | 8 | 2.32 | -6.6 | -7.06 | -2.49 | -1.68 | 1.78 | 1.1 |
| fatty acid binding | 11 | -1.61 | -1.59 | -2.07 | -1.15 | 1.3 | -1.68 | -1.56 |
| metal ion binding | 1381 | 1.03 | 1.02 | 1.04 | 1.03 | 1.11 | 1.05 | 1.01 |
| iron ion binding | 192 | -1.02 | -1.13 | -1.23 | -1.06 | -1.06 | 1.0 | -1.09 |
| copper ion binding | 20 | -1.87 | -1.23 | -1.09 | -1.21 | -1.25 | -1.59 | 1.03 |
| detection of calcium ion | 3 | -1.58 | -1.06 | -1.06 | 1.03 | 1.08 | -1.26 | -1.33 |
| sequestering of calcium ion | 1 | 1.04 | 1.04 | -1.04 | -1.09 | -1.16 | -1.14 | -1.1 |
| calmodulin binding | 30 | 1.18 | 1.09 | 1.27 | 1.05 | 1.23 | 1.34 | 1.23 |
| cytoskeletal regulatory protein binding | 1 | -1.55 | 1.24 | 2.16 | 1.01 | 1.73 | 1.49 | 1.71 |
| insulin-like growth factor binding | 3 | -1.07 | 1.13 | 1.1 | 1.18 | 1.32 | 1.03 | 1.12 |
| tropomyosin binding | 3 | 5.21 | -1.74 | 1.31 | -1.52 | 1.36 | 3.43 | -1.33 |
| ATP binding | 652 | 1.06 | 1.1 | 1.17 | 1.09 | 1.22 | 1.18 | 1.09 |
| GTP binding | 159 | 1.18 | 1.08 | 1.28 | 1.06 | 1.23 | 1.12 | 1.08 |
| macrolide binding | 6 | 1.43 | 1.09 | 1.09 | 1.21 | 1.26 | -1.11 | 1.17 |
| FK506 binding | 6 | 1.43 | 1.09 | 1.09 | 1.21 | 1.26 | -1.11 | 1.17 |
| sugar binding | 27 | -1.35 | -1.07 | 1.19 | -1.03 | -1.02 | -1.16 | 1.2 |
| galactose binding | 11 | -1.6 | -1.21 | 1.22 | -1.04 | -1.07 | -1.49 | -1.08 |
| mannose binding | 5 | -1.02 | 1.06 | -1.04 | 1.06 | -1.07 | -1.19 | -1.02 |
| glycosaminoglycan binding | 23 | -1.14 | -1.26 | -1.03 | -1.01 | -1.23 | -1.43 | -1.0 |
| folic acid binding | 4 | -1.96 | -1.02 | -2.85 | 1.48 | -1.35 | -1.15 | -2.07 |
| phospholipid binding | 112 | 1.08 | 1.08 | 1.36 | 1.02 | 1.26 | 1.28 | 1.26 |
| calcium-dependent phospholipid binding | 11 | 1.19 | -1.05 | 1.13 | 1.02 | 1.21 | 1.24 | 1.15 |
| 1-phosphatidylinositol binding | 1 | -1.06 | 1.07 | 1.15 | -1.0 | 1.23 | 1.29 | 1.44 |
| phosphatidylinositol-4,5-bisphosphate binding | 2 | -1.07 | 1.2 | 1.71 | 1.09 | 1.37 | 1.09 | 1.1 |
| phospholipid transporter activity | 14 | 1.01 | 1.02 | 1.0 | 1.1 | 1.38 | 1.01 | -1.05 |
| odorant binding | 110 | 1.09 | -1.06 | -1.12 | -1.07 | -1.13 | 1.0 | -1.08 |
| pheromone binding | 8 | 1.18 | 1.02 | -1.04 | 1.02 | -1.03 | -1.03 | -1.04 |
| protein ubiquitination | 41 | 1.11 | 1.13 | 1.39 | 1.14 | 1.24 | 1.15 | 1.15 |
| protein tag | 2 | 2.12 | -1.08 | -1.18 | 1.19 | 1.03 | -1.14 | 1.1 |
| ribosome | 169 | 1.2 | 1.05 | 1.14 | 1.12 | 1.17 | 1.28 | -1.17 |
| RNA modification guide activity | 2 | 1.38 | 1.09 | 1.14 | 1.35 | 1.29 | -1.02 | -1.04 |
| snRNA binding | 5 | 1.33 | 1.08 | 1.1 | 1.25 | 1.13 | 1.17 | -1.18 |
| cellular_component | 6965 | 1.08 | 1.01 | 1.05 | 1.02 | 1.09 | 1.09 | 1.03 |
| fibrinogen complex | 1 | -1.06 | -1.12 | -1.04 | 1.02 | 1.07 | -1.19 | -1.22 |
| proteinaceous extracellular matrix | 59 | 1.22 | -1.04 | 1.03 | -1.11 | 1.03 | 1.19 | -1.01 |
| collagen type IV | 2 | 1.15 | -1.7 | -1.07 | -1.48 | -1.42 | 2.42 | -1.33 |
| basement membrane | 10 | 1.55 | -1.19 | 1.13 | -1.32 | 1.1 | 2.13 | 1.42 |
| basal lamina | 3 | 2.42 | -1.01 | 1.11 | -1.14 | 1.47 | 4.1 | 1.43 |
| laminin-1 complex | 1 | 3.52 | 1.3 | 1.81 | 1.23 | 2.46 | 6.46 | 1.6 |
| integrin complex | 8 | -1.75 | 1.03 | 1.23 | -1.07 | 1.36 | -1.33 | 1.18 |
| interstitial matrix | 1 | -3.11 | 1.08 | 3.33 | -1.03 | 2.28 | -1.55 | -1.14 |
| extracellular space | 146 | -1.07 | -1.23 | -1.37 | -1.25 | -1.26 | -1.13 | 1.11 |
| larval serum protein complex | 5 | 1.15 | 1.01 | -1.14 | -1.06 | -1.16 | -1.08 | 1.13 |
| cell | 4600 | 1.14 | 1.06 | 1.13 | 1.06 | 1.16 | 1.15 | 1.06 |
| membrane fraction | 100 | -1.02 | -1.13 | -1.22 | -1.03 | 1.06 | -1.14 | -1.06 |
| soluble fraction | 6 | 1.33 | 1.04 | 1.25 | 1.02 | 1.26 | 1.47 | 1.19 |
| insoluble fraction | 103 | -1.03 | -1.12 | -1.22 | -1.02 | 1.06 | -1.11 | -1.05 |
| nucleus | 1578 | 1.19 | 1.1 | 1.2 | 1.12 | 1.23 | 1.17 | 1.1 |
| nuclear envelope | 72 | 1.34 | 1.09 | 1.32 | 1.13 | 1.35 | 1.29 | 1.14 |
| nuclear inner membrane | 7 | 1.5 | -1.14 | 1.37 | 1.22 | 1.72 | 1.02 | -1.06 |
| lamin filament | 2 | 1.28 | 1.06 | -1.0 | 1.32 | 1.15 | -1.17 | -1.1 |
| integral to nuclear inner membrane | 2 | -1.02 | -2.05 | 1.59 | 1.21 | 2.03 | -1.95 | -2.17 |
| nuclear outer membrane | 1 | 2.5 | -1.28 | -1.45 | -1.24 | 1.61 | 3.68 | 4.91 |
| nuclear envelope lumen | 2 | 1.16 | 1.03 | 1.1 | 1.15 | 1.11 | 1.57 | -1.14 |
| annulate lamellae | 1 | -1.39 | 1.32 | -1.25 | 1.42 | 1.03 | -1.29 | -1.2 |
| nuclear pore | 32 | 1.22 | 1.16 | 1.46 | 1.19 | 1.42 | 1.4 | 1.18 |
| Ran GTPase binding | 7 | -1.02 | 1.21 | 1.82 | 1.21 | 1.53 | 1.48 | 1.33 |
| cytoplasm | 2075 | 1.11 | 1.05 | 1.12 | 1.05 | 1.15 | 1.18 | 1.02 |
| nuclear lamina | 10 | 1.31 | 1.14 | 1.08 | 1.26 | 1.18 | 1.04 | 1.01 |
| nucleoplasm | 294 | 1.23 | 1.14 | 1.27 | 1.18 | 1.27 | 1.17 | 1.1 |
| nucleolar ribonuclease P complex | 1 | 1.84 | 1.03 | 1.1 | 1.42 | 1.17 | 1.05 | -1.14 |
| pre-replicative complex | 9 | -1.14 | 1.11 | 1.17 | 1.27 | 1.56 | -1.1 | -1.11 |
| replication fork | 18 | 1.16 | 1.11 | 1.19 | 1.26 | 1.39 | 1.09 | -1.16 |
| alpha DNA polymerase:primase complex | 4 | -1.1 | 1.07 | 1.15 | 1.07 | 1.18 | 1.06 | -1.25 |
| DNA replication factor C complex | 5 | 1.37 | 1.06 | 1.26 | 1.45 | 1.6 | 1.11 | -1.08 |
| PCNA complex | 2 | -1.29 | 1.09 | 1.33 | 1.22 | 1.3 | -1.6 | -1.28 |
| DNA replication factor A complex | 3 | 1.13 | 1.13 | 1.31 | 1.42 | 1.65 | 1.03 | -1.21 |
| nuclear origin of replication recognition complex | 7 | 1.06 | 1.15 | 1.09 | 1.19 | 1.24 | 1.01 | -1.08 |
| DNA-directed RNA polymerase II, core complex | 10 | 1.42 | 1.09 | 1.2 | 1.31 | 1.23 | 1.16 | 1.03 |
| DNA-directed RNA polymerase III complex | 8 | 1.27 | 1.21 | 1.22 | 1.2 | 1.28 | -1.02 | 1.11 |
| transcription factor complex | 83 | 1.26 | 1.14 | 1.3 | 1.14 | 1.22 | 1.14 | 1.12 |
| transcription factor TFIID complex | 23 | 1.34 | 1.16 | 1.26 | 1.14 | 1.29 | 1.17 | 1.16 |
| Ada2/Gcn5/Ada3 transcription activator complex | 8 | 1.4 | 1.19 | 1.36 | 1.24 | 1.39 | 1.07 | 1.22 |
| transcription factor TFIIA complex | 3 | 1.45 | -1.03 | 1.19 | 1.13 | 1.14 | 1.08 | -1.02 |
| transcription factor TFIIE complex | 2 | 1.86 | 1.06 | 1.16 | 1.23 | 1.25 | 1.2 | 1.16 |
| transcription factor TFIIF complex | 3 | 1.12 | 1.22 | 1.41 | 1.15 | 1.25 | 1.15 | 1.46 |
| holo TFIIH complex | 7 | 1.28 | 1.1 | 1.43 | 1.31 | 1.4 | 1.21 | 1.09 |
| chromatin assembly complex | 8 | 1.08 | -1.4 | -1.04 | -1.4 | 1.24 | 1.6 | 1.04 |
| anaphase-promoting complex | 10 | 1.4 | 1.11 | 1.22 | 1.13 | 1.2 | 1.12 | 1.1 |
| spliceosomal complex | 164 | 1.16 | 1.11 | 1.2 | 1.23 | 1.3 | 1.15 | -1.0 |
| U5 snRNP | 8 | 1.02 | 1.2 | 1.42 | 1.19 | 1.41 | 1.3 | 1.19 |
| U7 snRNP | 1 | 1.17 | -1.14 | -1.02 | 1.29 | 1.33 | -1.04 | -1.48 |
| U2-type spliceosomal complex | 1 | -1.28 | 1.17 | 1.49 | 1.3 | 1.15 | 1.37 | -1.07 |
| U1 snRNP | 9 | 1.08 | 1.24 | 1.47 | 1.25 | 1.39 | 1.33 | 1.11 |
| U2 snRNP | 13 | 1.25 | 1.19 | 1.29 | 1.24 | 1.28 | 1.19 | 1.05 |
| U4 snRNP | 2 | 1.38 | 1.31 | 1.34 | 1.36 | 1.44 | 1.2 | 1.04 |
| U6 snRNP | 9 | 1.23 | -1.02 | 1.09 | 1.34 | 1.26 | -1.1 | -1.27 |
| U11 snRNP | 2 | 1.15 | 1.06 | 1.1 | 1.32 | 1.06 | 1.17 | -1.28 |
| U12 snRNP | 1 | 1.7 | -1.05 | -1.07 | -1.09 | -1.18 | 1.33 | 1.14 |
| chromosome | 301 | 1.18 | 1.13 | 1.24 | 1.14 | 1.29 | 1.19 | 1.11 |
| polytene chromosome | 101 | 1.23 | 1.19 | 1.33 | 1.13 | 1.37 | 1.36 | 1.27 |
| polytene chromosome chromocenter | 12 | 1.15 | 1.01 | 1.1 | 1.06 | 1.4 | 1.13 | 1.11 |
| polytene chromosome puff | 21 | 1.18 | 1.24 | 1.27 | 1.17 | 1.25 | 1.41 | 1.23 |
| polytene chromosome band | 8 | 1.19 | 1.12 | 1.3 | 1.15 | 1.48 | 1.11 | 1.11 |
| polytene chromosome interband | 20 | 1.23 | 1.21 | 1.27 | 1.2 | 1.3 | 1.27 | 1.15 |
| nuclear euchromatin | 4 | 1.52 | 1.39 | 1.51 | 1.2 | 1.41 | 1.55 | 1.43 |
| nuclear heterochromatin | 9 | 1.18 | 1.12 | 1.33 | 1.09 | 1.48 | 1.23 | 1.27 |
| centromeric heterochromatin | 10 | 1.2 | 1.1 | 1.3 | 1.02 | 1.42 | 1.45 | 1.42 |
| beta-heterochromatin | 1 | 1.4 | 1.27 | 1.49 | -1.01 | 1.2 | 1.7 | 1.58 |
| alpha-heterochromatin | 1 | 1.02 | 1.05 | 1.18 | 1.16 | 1.33 | 1.09 | 1.01 |
| nuclear telomeric heterochromatin | 1 | -1.17 | 1.26 | 1.25 | 1.08 | 2.11 | -1.1 | 1.25 |
| intercalary heterochromatin | 6 | 1.31 | 1.18 | 1.31 | 1.11 | 1.44 | 1.27 | 1.36 |
| nucleolus | 70 | 1.19 | 1.16 | 1.25 | 1.22 | 1.26 | 1.08 | 1.1 |
| small nucleolar ribonucleoprotein complex | 5 | 1.37 | 1.06 | 1.15 | 1.38 | 1.26 | -1.06 | -1.08 |
| DNA-directed RNA polymerase I complex | 6 | 1.23 | 1.16 | 1.31 | 1.23 | 1.28 | 1.23 | 1.09 |
| mitochondrial envelope | 204 | 1.05 | 1.07 | 1.15 | 1.05 | 1.1 | 1.39 | -1.2 |
| mitochondrial outer membrane | 23 | 1.13 | -1.01 | 1.18 | 1.12 | 1.17 | 1.29 | -1.07 |
| mitochondrial outer membrane translocase complex | 7 | 1.18 | 1.07 | 1.36 | 1.14 | 1.17 | 1.44 | -1.07 |
| mitochondrial inner membrane | 153 | 1.04 | 1.06 | 1.17 | 1.05 | 1.09 | 1.4 | -1.23 |
| mitochondrial inner membrane presequence translocase complex | 11 | 1.06 | 1.12 | 1.2 | 1.14 | 1.14 | 1.28 | -1.15 |
| mitochondrial respiratory chain | 68 | 1.11 | 1.0 | 1.19 | 1.01 | 1.07 | 1.57 | -1.31 |
| mitochondrial respiratory chain complex I | 37 | 1.15 | 1.04 | 1.27 | 1.07 | 1.11 | 1.72 | -1.29 |
| mitochondrial respiratory chain complex II | 6 | 1.02 | 1.03 | 1.28 | -1.03 | -1.0 | 1.38 | -1.21 |
| mitochondrial respiratory chain complex III | 11 | -1.13 | 1.03 | 1.1 | 1.04 | -1.01 | 1.3 | -1.48 |
| mitochondrial respiratory chain complex IV | 13 | 1.04 | -1.01 | 1.2 | -1.0 | 1.14 | 1.38 | -1.29 |
| mitochondrial proton-transporting ATP synthase complex | 18 | 1.31 | -1.01 | 1.22 | -1.03 | 1.11 | 1.68 | -1.22 |
| proton-transporting ATP synthase complex, coupling factor F(o) | 9 | 1.19 | 1.04 | 1.31 | 1.01 | 1.22 | 1.62 | -1.14 |
| mitochondrial proton-transporting ATP synthase, central stalk | 1 | 1.03 | 1.03 | 1.17 | 1.01 | 1.11 | 1.58 | -1.38 |
| mitochondrial intermembrane space | 11 | 1.06 | 1.14 | 1.24 | 1.17 | 1.16 | 1.38 | -1.17 |
| mitochondrial matrix | 144 | 1.1 | 1.08 | 1.17 | 1.11 | 1.17 | 1.48 | -1.3 |
| gamma DNA polymerase complex | 2 | -1.07 | 1.23 | 1.2 | 1.04 | 1.03 | 1.03 | -1.12 |
| mitochondrial ribosome | 72 | 1.04 | 1.07 | 1.23 | 1.29 | 1.25 | 1.45 | -1.55 |
| mitochondrial large ribosomal subunit | 44 | 1.03 | 1.05 | 1.24 | 1.3 | 1.28 | 1.46 | -1.6 |
| mitochondrial small ribosomal subunit | 30 | 1.04 | 1.09 | 1.24 | 1.27 | 1.23 | 1.47 | -1.48 |
| lysosomal membrane | 4 | 1.09 | -1.11 | -1.38 | 1.13 | 1.63 | 2.55 | 1.04 |
| secondary lysosome | 1 | 1.17 | -2.03 | 2.67 | -1.12 | -2.24 | 1.02 | -1.06 |
| endosome | 44 | -1.01 | 1.1 | 1.4 | 1.04 | 1.31 | 1.27 | 1.37 |
| early endosome | 19 | -1.03 | 1.08 | 1.36 | 1.01 | 1.16 | 1.2 | 1.43 |
| late endosome | 17 | -1.09 | 1.11 | 1.22 | 1.07 | 1.21 | 1.3 | 1.36 |
| multivesicular body | 6 | -1.01 | 1.0 | 1.49 | 1.07 | 1.26 | 1.31 | 1.59 |
| vacuolar membrane | 30 | -1.55 | -1.0 | 1.28 | -1.14 | -1.08 | 1.41 | -1.12 |
| integral to peroxisomal membrane | 5 | 1.37 | 1.29 | -1.51 | 1.15 | 1.08 | -1.33 | -1.13 |
| protein targeting to peroxisome | 3 | 1.5 | 1.17 | -1.6 | 1.44 | 1.46 | -2.19 | -1.64 |
| peroxisomal part | 14 | 1.87 | 1.14 | -1.32 | 1.28 | 1.36 | -1.33 | -1.09 |
| Sec61 translocon complex | 8 | -1.51 | 1.3 | -1.14 | -1.0 | -1.28 | -1.7 | 1.22 |
| signal recognition particle receptor complex | 4 | -1.91 | 1.42 | -1.08 | 1.06 | -1.35 | -1.79 | 1.42 |
| signal recognition particle, endoplasmic reticulum targeting | 9 | -1.25 | 1.37 | 1.01 | 1.14 | -1.09 | -1.57 | 1.26 |
| signal peptidase complex | 3 | 1.12 | -1.0 | 1.0 | -1.02 | -1.02 | -1.01 | 1.75 |
| endoplasmic reticulum lumen | 9 | 1.27 | -1.04 | 1.01 | 1.06 | 1.11 | -1.04 | 1.39 |
| endoplasmic reticulum membrane | 64 | -1.15 | 1.18 | 1.16 | 1.11 | 1.13 | -1.17 | 1.12 |
| smooth endoplasmic reticulum | 5 | 2.12 | -1.15 | 1.68 | -1.13 | 1.04 | 1.12 | 1.23 |
| rough endoplasmic reticulum | 15 | -1.44 | 1.23 | -1.0 | 1.01 | -1.26 | -1.5 | 1.29 |
| microsome | 85 | -1.05 | -1.16 | -1.3 | -1.04 | 1.03 | -1.24 | -1.12 |
| endoplasmic reticulum-Golgi intermediate compartment | 1 | -1.08 | 1.49 | -1.64 | 1.02 | -1.4 | -2.11 | -1.11 |
| Golgi apparatus | 138 | 1.04 | 1.15 | 1.18 | 1.1 | 1.19 | 1.13 | 1.3 |
| Golgi stack | 31 | 1.11 | 1.08 | 1.07 | 1.04 | 1.18 | 1.12 | 1.47 |
| Golgi-associated vesicle | 20 | -1.22 | 1.18 | 1.09 | 1.15 | 1.06 | 1.03 | 1.23 |
| COPI vesicle coat | 9 | -1.24 | 1.16 | -1.05 | 1.14 | -1.11 | -1.1 | 1.28 |
| COPI-coated vesicle | 9 | -1.24 | 1.16 | -1.05 | 1.14 | -1.11 | -1.1 | 1.28 |
| ER to Golgi transport vesicle | 4 | 1.31 | 1.22 | 1.29 | 1.04 | 1.28 | 1.43 | 2.03 |
| cis-Golgi network | 9 | 1.04 | 1.19 | 1.21 | 1.18 | 1.15 | 1.12 | 1.14 |
| trans-Golgi network | 6 | -1.72 | 1.13 | 1.32 | 1.15 | 1.37 | 1.03 | 1.11 |
| transport vesicle | 15 | 1.23 | 1.06 | 1.2 | -1.01 | 1.17 | 1.53 | 1.45 |
| secretory granule | 4 | -1.19 | 1.11 | 1.32 | 1.05 | 1.1 | -1.29 | 1.16 |
| transport vesicle membrane | 4 | -1.16 | 1.25 | 1.59 | 1.05 | 1.44 | 1.76 | 1.54 |
| endocytic vesicle | 10 | -1.1 | -1.41 | -1.31 | -1.37 | 1.06 | 1.46 | 1.02 |
| lipid particle | 232 | 1.17 | -1.07 | -1.13 | -1.09 | 1.05 | 1.16 | -1.03 |
| centrosome | 62 | 1.25 | 1.07 | 1.27 | 1.05 | 1.21 | 1.28 | 1.12 |
| centriole | 10 | 1.09 | 1.08 | 1.02 | 1.1 | 1.08 | -1.01 | -1.07 |
| microtubule organizing center | 72 | 1.25 | 1.07 | 1.25 | 1.06 | 1.21 | 1.27 | 1.11 |
| aster | 7 | -1.26 | 1.17 | 1.31 | 1.12 | 1.35 | -1.04 | 1.04 |
| spindle | 81 | 1.21 | 1.11 | 1.28 | 1.11 | 1.35 | 1.24 | 1.13 |
| actomyosin contractile ring | 4 | -1.06 | 1.15 | 1.51 | 1.14 | 1.77 | 1.47 | 1.16 |
| polar microtubule | 2 | -1.12 | 1.17 | 1.2 | 1.12 | 1.31 | 1.44 | 1.21 |
| kinetochore microtubule | 9 | -1.0 | 1.13 | 1.07 | 1.14 | 1.19 | 1.02 | 1.02 |
| chaperonin-containing T-complex | 8 | -1.27 | 1.13 | 1.1 | 1.15 | -1.02 | -1.22 | -1.18 |
| fatty acid synthase complex | 1 | 1.39 | -1.11 | 1.17 | 1.12 | 1.04 | 1.29 | -1.0 |
| proteasome regulatory particle | 24 | 1.35 | 1.08 | 1.26 | 1.17 | 1.2 | 1.06 | 1.07 |
| proteasome core complex | 23 | 1.34 | -1.02 | 1.1 | 1.11 | 1.12 | -1.15 | -1.1 |
| polysome | 1 | 2.2 | 1.38 | 1.27 | 1.25 | 1.7 | 1.45 | 1.27 |
| mRNA cap binding complex | 2 | 1.35 | 1.18 | 1.45 | 1.11 | 1.5 | 1.76 | 1.35 |
| nuclear cap binding complex | 2 | 1.33 | 1.12 | 1.23 | 1.21 | 1.34 | 1.16 | 1.07 |
| mRNA cleavage and polyadenylation specificity factor complex | 7 | 1.3 | 1.31 | 1.42 | 1.47 | 1.64 | 1.34 | 1.15 |
| mRNA cleavage stimulating factor complex | 4 | 1.19 | 1.15 | 1.23 | 1.16 | 1.21 | 1.09 | 1.06 |
| mRNA cleavage factor complex | 13 | 1.19 | 1.28 | 1.36 | 1.32 | 1.43 | 1.3 | 1.14 |
| eukaryotic translation initiation factor 2 complex | 3 | 1.23 | 1.22 | 1.28 | 1.08 | 1.12 | 1.46 | 1.17 |
| eukaryotic translation initiation factor 2B complex | 6 | 1.27 | 1.3 | 1.06 | 1.17 | 1.06 | 1.14 | 1.04 |
| eukaryotic translation initiation factor 3 complex | 16 | 1.44 | 1.14 | 1.33 | 1.03 | 1.2 | 1.61 | 1.17 |
| eukaryotic translation elongation factor 1 complex | 5 | 2.72 | -1.02 | 1.29 | -1.13 | 1.23 | 1.37 | 1.32 |
| nascent polypeptide-associated complex | 3 | 1.55 | -1.11 | 1.12 | 1.0 | 1.05 | 1.16 | 1.07 |
| cytoskeleton | 548 | 1.09 | 1.04 | 1.16 | 1.03 | 1.2 | 1.15 | 1.06 |
| muscle myosin complex | 5 | 4.08 | -1.62 | 1.08 | -1.52 | 1.03 | 2.86 | -1.26 |
| troponin complex | 2 | 6.95 | -2.33 | -1.12 | -1.86 | 1.08 | 3.33 | -1.76 |
| muscle thin filament tropomyosin | 2 | 3.23 | -1.38 | 1.31 | -1.15 | 1.21 | 2.24 | -1.26 |
| striated muscle myosin thick filament | 3 | 2.91 | -1.49 | 1.05 | -1.32 | 1.04 | 2.39 | -1.29 |
| striated muscle thin filament | 5 | 3.24 | -1.5 | 1.09 | -1.38 | 1.25 | 2.3 | -1.28 |
| dynactin complex | 10 | 1.11 | 1.07 | 1.24 | 1.19 | 1.25 | 1.09 | -1.06 |
| kinesin complex | 29 | 1.08 | 1.11 | 1.16 | 1.1 | 1.2 | 1.14 | 1.1 |
| minus-end kinesin complex | 1 | -1.34 | -1.01 | 1.11 | 1.33 | 1.45 | -1.15 | -1.3 |
| plus-end kinesin complex | 3 | 1.17 | 1.1 | 1.09 | 1.07 | 1.19 | 1.04 | -1.14 |
| microtubule | 54 | 1.03 | 1.09 | 1.21 | 1.07 | 1.23 | 1.11 | 1.08 |
| microtubule associated complex | 344 | 1.12 | 1.03 | 1.11 | 1.03 | 1.19 | 1.17 | 1.06 |
| spindle microtubule | 24 | 1.15 | 1.11 | 1.33 | 1.06 | 1.29 | 1.35 | 1.18 |
| axonemal microtubule | 1 | -1.31 | -1.04 | -1.2 | -1.01 | -1.13 | -1.03 | -1.15 |
| nuclear microtubule | 1 | 1.51 | 1.15 | 1.47 | 1.1 | 1.35 | 1.02 | -1.14 |
| cytoplasmic microtubule | 9 | -1.07 | 1.19 | 1.41 | 1.03 | 1.48 | 1.12 | 1.24 |
| actin filament | 26 | -1.06 | -1.0 | 1.14 | -1.03 | 1.19 | 1.09 | 1.05 |
| Arp2/3 protein complex | 5 | -3.04 | -1.69 | -1.35 | 1.45 | 1.29 | -2.9 | -1.36 |
| sodium:potassium-exchanging ATPase complex | 8 | 1.13 | -1.11 | 1.22 | -1.35 | -1.0 | 1.53 | 1.06 |
| voltage-gated calcium channel complex | 4 | 1.36 | -1.08 | -1.02 | 1.02 | -1.08 | 1.17 | -1.12 |
| acetylcholine-gated channel complex | 9 | -1.07 | 1.01 | -1.02 | -1.03 | -1.09 | -1.06 | -1.03 |
| insulin receptor complex | 1 | 1.01 | 1.43 | 1.57 | 1.5 | 2.27 | 1.72 | 1.64 |
| caveola | 1 | 3.68 | -1.06 | -1.03 | 1.16 | 1.0 | 1.99 | -1.04 |
| microvillus | 5 | 1.36 | 1.2 | 1.61 | -1.02 | 1.17 | 1.09 | 1.29 |
| brush border | 3 | -22.7 | -1.04 | -2.81 | 1.01 | -1.94 | 1.91 | -2.7 |
| coated pit | 6 | -1.26 | 1.18 | 1.24 | 1.12 | 1.21 | 1.33 | 1.27 |
| AP-type membrane coat adaptor complex | 9 | -1.11 | 1.2 | 1.31 | 1.16 | 1.2 | 1.14 | 1.2 |
| AP-1 adaptor complex | 1 | -1.46 | 1.16 | 1.32 | 1.12 | 1.05 | 1.44 | 1.08 |
| cell-cell junction | 66 | 1.52 | -1.07 | 1.19 | -1.11 | 1.18 | 1.85 | 1.41 |
| adherens junction | 58 | 1.23 | 1.15 | 1.4 | 1.04 | 1.4 | 1.35 | 1.31 |
| cell-cell adherens junction | 13 | 1.51 | 1.27 | 1.67 | 1.06 | 1.61 | 1.75 | 1.74 |
| spot adherens junction | 4 | 1.12 | 1.24 | 1.44 | 1.07 | 1.52 | 1.53 | 1.48 |
| zonula adherens | 9 | 1.2 | 1.26 | 1.62 | 1.08 | 1.63 | 1.49 | 1.6 |
| nephrocyte diaphragm | 3 | -3.44 | 1.01 | 1.01 | 1.15 | 1.01 | -2.84 | -1.29 |
| septate junction | 22 | 2.96 | -1.28 | 1.21 | -1.34 | 1.26 | 3.68 | 1.76 |
| pleated septate junction | 3 | 3.11 | -1.4 | 1.27 | -1.39 | 1.24 | 4.46 | 1.69 |
| gap junction | 8 | -1.22 | -1.27 | -1.23 | -1.25 | -1.39 | 1.09 | 1.54 |
| tight junction | 4 | 1.14 | 1.05 | 1.46 | 1.03 | 1.1 | 1.55 | 1.04 |
| cell-substrate adherens junction | 21 | 1.3 | 1.08 | 1.33 | 1.01 | 1.35 | 1.25 | 1.25 |
| focal adhesion | 15 | 1.13 | 1.11 | 1.37 | 1.02 | 1.47 | 1.18 | 1.23 |
| connecting hemi-adherens junction | 1 | 1.1 | 1.42 | 2.42 | 1.36 | 2.63 | 2.12 | 3.14 |
| muscle tendon junction | 5 | 2.04 | -1.05 | 1.1 | -1.07 | -1.09 | 1.36 | 1.08 |
| cilium | 6 | 1.05 | 1.08 | 1.01 | 1.11 | -1.01 | -1.05 | 1.1 |
| microtubule basal body | 4 | 1.06 | -1.02 | 1.03 | 1.0 | -1.01 | 1.11 | -1.03 |
| cell cortex | 86 | -1.07 | 1.18 | 1.41 | 1.09 | 1.4 | 1.22 | 1.2 |
| septin ring | 5 | -1.56 | 1.08 | 1.09 | 1.16 | 1.34 | -1.29 | 1.06 |
| phosphatidylinositol 3-kinase complex | 4 | -1.6 | 1.29 | 1.4 | 1.19 | 1.74 | -1.12 | -1.04 |
| 1-phosphatidylinositol-4-phosphate 3-kinase, class IA complex | 2 | -1.54 | 1.47 | 1.35 | 1.15 | 1.83 | 1.07 | 1.48 |
| 6-phosphofructokinase complex | 1 | 2.63 | -1.22 | -1.22 | 1.02 | -1.12 | 5.11 | 1.3 |
| mitochondrial alpha-ketoglutarate dehydrogenase complex | 1 | -1.4 | -1.14 | 2.27 | -1.25 | -1.48 | 1.94 | -1.5 |
| cAMP-dependent protein kinase complex | 6 | 1.3 | 1.06 | 1.3 | -1.12 | 1.18 | 1.98 | 1.03 |
| calcium- and calmodulin-dependent protein kinase complex | 2 | 1.81 | 1.24 | 1.48 | 1.18 | 1.44 | 2.31 | 1.51 |
| calcineurin complex | 5 | 1.63 | -1.01 | 1.24 | 1.04 | 1.14 | 1.3 | 1.06 |
| protein kinase CK2 complex | 5 | -1.01 | -1.08 | 1.03 | -1.07 | 1.06 | -1.06 | -1.08 |
| DNA-dependent protein kinase-DNA ligase 4 complex | 2 | -1.03 | 1.29 | 1.58 | 1.29 | 1.51 | -1.21 | 1.27 |
| glycine cleavage complex | 4 | 1.1 | -1.07 | 1.31 | -1.13 | -1.08 | 2.33 | -1.17 |
| mitochondrial isocitrate dehydrogenase complex (NAD+) | 1 | -1.07 | 1.16 | -1.05 | -1.13 | 1.15 | 1.71 | 1.06 |
| phosphorylase kinase complex | 3 | 1.8 | 1.28 | 1.21 | 1.07 | 1.13 | 2.26 | 1.63 |
| protein farnesyltransferase complex | 1 | -1.83 | -1.28 | -1.22 | 1.05 | -1.01 | -1.04 | 2.75 |
| mitochondrial pyruvate dehydrogenase complex | 1 | 1.23 | 1.19 | 1.96 | -1.07 | 1.35 | 3.33 | 1.11 |
| Rab-protein geranylgeranyltransferase complex | 4 | -1.3 | 1.2 | 1.28 | 1.18 | 1.3 | -1.38 | -1.0 |
| ribonucleoside-diphosphate reductase complex | 2 | -2.27 | -1.06 | 1.44 | 1.36 | 1.87 | -1.61 | -1.54 |
| carbohydrate metabolic process | 349 | -1.3 | -1.19 | -1.48 | -1.16 | -1.22 | -1.28 | 1.04 |
| polysaccharide metabolic process | 131 | -1.49 | -1.56 | -1.71 | -1.43 | -1.46 | -1.36 | 1.27 |
| glycogen metabolic process | 9 | 2.15 | 1.01 | -1.12 | 1.0 | 1.05 | 2.73 | 1.34 |
| glycogen biosynthetic process | 5 | 2.74 | -1.19 | -1.42 | 1.04 | 1.07 | 2.78 | 1.29 |
| glycogen catabolic process | 1 | 2.98 | -1.08 | 1.5 | -1.31 | 1.12 | 4.86 | 1.33 |
| disaccharide metabolic process | 5 | 2.21 | -1.79 | -2.13 | -1.71 | -1.22 | 2.7 | -1.01 |
| trehalose metabolic process | 5 | 2.21 | -1.79 | -2.13 | -1.71 | -1.22 | 2.7 | -1.01 |
| trehalose biosynthetic process | 3 | 3.57 | -2.83 | -2.37 | -2.36 | -1.16 | 5.45 | -1.46 |
| monosaccharide metabolic process | 74 | -1.39 | -1.01 | -1.54 | 1.02 | -1.04 | -1.38 | -1.41 |
| fructose metabolic process | 1 | 1.09 | -1.34 | 2.59 | -1.04 | 1.14 | 3.38 | -1.06 |
| fructose 2,6-bisphosphate metabolic process | 1 | 1.09 | -1.34 | 2.59 | -1.04 | 1.14 | 3.38 | -1.06 |
| fucose metabolic process | 4 | -1.24 | 1.16 | -1.09 | -1.11 | 1.15 | -1.73 | 2.16 |
| L-fucose biosynthetic process | 2 | -1.67 | 1.21 | 1.13 | 1.24 | 1.36 | -1.89 | -1.14 |
| glucose metabolic process | 42 | -1.0 | -1.08 | -1.42 | -1.06 | -1.11 | 1.05 | -1.04 |
| glucose catabolic process | 33 | 1.52 | 1.06 | -1.18 | -1.03 | -1.06 | 1.37 | 1.2 |
| galactose metabolic process | 1 | -2.18 | 1.44 | -1.11 | -1.39 | 1.11 | -2.6 | 2.04 |
| mannose metabolic process | 11 | -9.63 | -1.07 | -4.72 | 1.44 | -1.08 | -9.35 | -13.32 |
| D-ribose metabolic process | 2 | -1.21 | 1.07 | -1.3 | 1.1 | -1.27 | -1.38 | -1.12 |
| inositol metabolic process | 2 | 2.7 | -2.43 | -2.03 | -2.2 | -1.38 | 3.25 | -1.23 |
| inositol biosynthetic process | 1 | 1.86 | -1.46 | -1.12 | -1.51 | 1.13 | 1.83 | 1.71 |
| aminoglycan metabolic process | 120 | -1.64 | -1.63 | -1.78 | -1.48 | -1.53 | -1.5 | 1.27 |
| aminoglycan biosynthetic process | 15 | 1.36 | 1.11 | 1.12 | 1.03 | 1.14 | 1.41 | 1.3 |
| glycosaminoglycan biosynthetic process | 11 | 1.22 | 1.23 | 1.19 | 1.13 | 1.29 | 1.22 | 1.28 |
| aminoglycan catabolic process | 29 | -1.22 | -1.95 | -2.17 | -1.68 | -1.71 | -1.1 | 1.4 |
| glycosaminoglycan catabolic process | 12 | -1.13 | -1.64 | -1.19 | -1.04 | -1.82 | -1.69 | -1.29 |
| chitin metabolic process | 96 | -1.91 | -1.76 | -2.05 | -1.64 | -1.62 | -1.64 | 1.34 |
| chitin biosynthetic process | 4 | 1.85 | -1.21 | -1.08 | -1.24 | -1.23 | 2.11 | 1.33 |
| chitin catabolic process | 17 | -1.28 | -2.2 | -3.31 | -2.38 | -1.64 | 1.24 | 2.14 |
| cuticle chitin metabolic process | 7 | -1.6 | -1.5 | -2.17 | -1.7 | -1.06 | 1.36 | 1.67 |
| cuticle chitin biosynthetic process | 3 | 1.16 | -1.14 | 1.05 | -1.21 | -1.11 | 2.09 | 1.42 |
| cuticle chitin catabolic process | 4 | -2.55 | -1.85 | -4.04 | -2.19 | -1.03 | -1.02 | 1.9 |
| amino sugar metabolic process | 9 | 1.34 | 1.24 | 1.13 | 1.09 | 1.26 | 1.2 | 1.12 |
| glucosamine metabolic process | 8 | 1.39 | 1.28 | 1.16 | 1.11 | 1.3 | 1.22 | 1.15 |
| N-acetylglucosamine metabolic process | 8 | 1.39 | 1.28 | 1.16 | 1.11 | 1.3 | 1.22 | 1.15 |
| N-acetylneuraminate metabolic process | 1 | 1.0 | -1.04 | -1.09 | -1.07 | -1.02 | 1.03 | -1.14 |
| CMP-N-acetylneuraminate biosynthetic process | 1 | 1.0 | -1.04 | -1.09 | -1.07 | -1.02 | 1.03 | -1.14 |
| alcohol metabolic process | 137 | -1.15 | -1.03 | -1.35 | 1.01 | -1.01 | -1.18 | -1.15 |
| ethanol metabolic process | 2 | 1.95 | -1.28 | -2.91 | -1.89 | 1.06 | 1.72 | 1.15 |
| ethanol oxidation | 2 | 1.95 | -1.28 | -2.91 | -1.89 | 1.06 | 1.72 | 1.15 |
| glycerol metabolic process | 16 | -1.18 | 1.1 | -1.05 | 1.11 | 1.01 | -1.33 | -1.11 |
| glycerol-3-phosphate metabolic process | 10 | 1.18 | -1.01 | -1.04 | 1.17 | 1.39 | 1.07 | -1.25 |
| cellular glucan metabolic process | 9 | 2.15 | 1.01 | -1.12 | 1.0 | 1.05 | 2.73 | 1.34 |
| cellular aldehyde metabolic process | 8 | 1.58 | -1.07 | -1.41 | -1.33 | -1.03 | 1.21 | 1.04 |
| organic acid metabolic process | 239 | -1.01 | -1.01 | -1.15 | -1.03 | 1.0 | 1.06 | -1.05 |
| acetyl-CoA metabolic process | 39 | -1.0 | 1.09 | 1.25 | -1.05 | 1.07 | 1.5 | -1.03 |
| acetyl-CoA biosynthetic process | 2 | -1.03 | 1.03 | 1.25 | -1.03 | 1.02 | 1.46 | -1.0 |
| acetyl-CoA biosynthetic process from pyruvate | 1 | 1.23 | 1.19 | 1.96 | -1.07 | 1.35 | 3.33 | 1.11 |
| pyruvate metabolic process | 10 | 1.06 | 1.22 | 1.23 | -1.11 | 1.04 | 1.51 | 1.12 |
| generation of precursor metabolites and energy | 140 | 1.11 | 1.05 | 1.13 | 1.0 | 1.09 | 1.53 | -1.12 |
| gluconeogenesis | 4 | 1.09 | -1.28 | -1.21 | -1.39 | 1.08 | 1.63 | -1.02 |
| glycolysis | 25 | 1.37 | 1.03 | -1.05 | -1.05 | -1.07 | 1.43 | 1.11 |
| glyoxylate cycle | 1 | 1.5 | 1.55 | -1.65 | -1.56 | -1.01 | -1.09 | 1.26 |
| pentose-phosphate shunt | 7 | 2.13 | 1.17 | -1.85 | 1.03 | -1.09 | 1.03 | 1.53 |
| tricarboxylic acid cycle | 36 | -1.0 | 1.09 | 1.26 | -1.05 | 1.07 | 1.52 | -1.04 |
| citrate metabolic process | 2 | -1.17 | -1.01 | -1.18 | 1.04 | -1.26 | -1.29 | -1.1 |
| 2-oxoglutarate metabolic process | 2 | -1.13 | -1.34 | 1.15 | -1.41 | 1.89 | 2.28 | -2.14 |
| succinate metabolic process | 2 | -1.04 | 1.09 | 1.19 | 1.01 | 1.02 | 1.36 | -1.32 |
| fumarate metabolic process | 6 | -1.06 | -1.08 | 1.01 | -1.19 | 1.09 | 1.49 | -1.51 |
| malate metabolic process | 6 | -1.07 | 1.16 | 1.1 | -1.34 | -1.06 | 1.05 | 1.38 |
| isocitrate metabolic process | 1 | 1.5 | 1.55 | -1.65 | -1.56 | -1.01 | -1.09 | 1.26 |
| regulation of carbohydrate metabolic process | 3 | -1.56 | -1.05 | -1.43 | -1.32 | 1.05 | 1.6 | 2.11 |
| energy reserve metabolic process | 9 | 2.15 | 1.01 | -1.12 | 1.0 | 1.05 | 2.73 | 1.34 |
| NADH oxidation | 1 | -1.32 | -1.9 | 1.3 | -2.14 | 3.35 | 4.87 | -4.48 |
| acetaldehyde metabolic process | 2 | 2.78 | -1.16 | -2.38 | -2.31 | -1.44 | 1.4 | 1.22 |
| oxidation-reduction process | 538 | 1.03 | -1.05 | -1.16 | -1.04 | -1.02 | 1.09 | -1.04 |
| oxidoreductase activity | 612 | 1.05 | -1.06 | -1.18 | -1.04 | -1.04 | 1.03 | -1.05 |
| respiratory electron transport chain | 63 | 1.04 | 1.03 | 1.18 | 1.06 | 1.12 | 1.53 | -1.32 |
| oxidative phosphorylation | 65 | 1.08 | 1.05 | 1.19 | 1.05 | 1.1 | 1.53 | -1.32 |
| mitochondrial electron transport, NADH to ubiquinone | 31 | 1.15 | 1.03 | 1.29 | 1.07 | 1.13 | 1.74 | -1.29 |
| mitochondrial electron transport, succinate to ubiquinone | 3 | -1.1 | 1.07 | 1.36 | -1.03 | -1.0 | 1.57 | -1.31 |
| mitochondrial electron transport, ubiquinol to cytochrome c | 12 | -1.09 | 1.04 | 1.06 | 1.05 | -1.0 | 1.32 | -1.48 |
| mitochondrial electron transport, cytochrome c to oxygen | 11 | 1.03 | -1.01 | 1.21 | 1.02 | 1.15 | 1.4 | -1.32 |
| cell redox homeostasis | 50 | 1.14 | 1.05 | 1.09 | 1.13 | 1.1 | 1.12 | -1.05 |
| electron transport chain | 67 | -1.04 | 1.03 | 1.17 | 1.06 | 1.11 | 1.45 | -1.31 |
| glycerophosphate shuttle | 2 | 1.11 | -1.19 | -1.41 | 1.22 | 2.52 | 1.34 | -1.21 |
| glutathione metabolic process | 4 | -2.31 | 1.0 | -1.94 | 1.16 | 1.13 | -1.72 | 1.13 |
| nucleobase-containing compound metabolic process | 986 | 1.18 | 1.11 | 1.19 | 1.14 | 1.24 | 1.16 | 1.06 |
| regulation of nucleotide metabolic process | 51 | 1.09 | 1.14 | 1.33 | 1.05 | 1.25 | 1.28 | 1.17 |
| purine base metabolic process | 6 | 1.48 | 1.02 | -1.13 | -1.0 | -1.02 | 1.22 | -1.12 |
| purine nucleotide metabolic process | 118 | 1.13 | 1.02 | 1.13 | 1.0 | 1.06 | 1.27 | -1.04 |
| purine nucleoside metabolic process | 12 | 1.18 | 1.15 | -1.0 | 1.18 | 1.26 | -1.09 | -1.09 |
| purine nucleotide biosynthetic process | 67 | 1.06 | 1.03 | 1.1 | 1.0 | 1.03 | 1.3 | -1.07 |
| nucleoside diphosphate phosphorylation | 5 | 1.01 | 1.09 | 1.35 | 1.24 | 1.35 | -1.0 | -1.14 |
| purine ribonucleoside salvage | 4 | 1.03 | 1.11 | -1.11 | 1.23 | 1.2 | 1.03 | -1.31 |
| adenine salvage | 1 | 1.17 | 1.37 | 1.23 | 1.24 | 1.3 | 1.33 | -1.5 |
| cAMP biosynthetic process | 6 | 1.38 | -1.01 | 1.27 | 1.01 | 1.07 | 1.29 | 1.16 |
| ADP biosynthetic process | 2 | -1.29 | -1.01 | -1.04 | 1.37 | 1.42 | -2.05 | -1.57 |
| GMP biosynthetic process | 2 | 1.15 | 1.21 | 1.23 | -1.01 | 1.16 | 1.11 | 2.05 |
| cGMP biosynthetic process | 10 | -1.15 | -1.02 | -1.07 | 1.01 | -1.08 | -1.14 | -1.12 |
| GTP biosynthetic process | 4 | 1.0 | 1.09 | 1.36 | 1.22 | 1.36 | 1.06 | -1.14 |
| GTP catabolic process | 37 | 1.17 | 1.04 | 1.14 | 1.08 | 1.12 | 1.08 | 1.0 |
| IMP biosynthetic process | 7 | 1.14 | 1.03 | -1.12 | -1.04 | -1.13 | 1.95 | -1.01 |
| 'de novo' IMP biosynthetic process | 6 | 1.13 | 1.02 | -1.2 | -1.03 | -1.2 | 1.78 | -1.03 |
| purine nucleotide catabolic process | 40 | 1.18 | 1.04 | 1.15 | 1.09 | 1.13 | 1.08 | 1.01 |
| ATP catabolic process | 2 | 1.12 | 1.21 | 1.11 | 1.15 | 1.2 | 1.05 | 1.15 |
| pyrimidine base metabolic process | 9 | 1.42 | 1.06 | -1.02 | 1.04 | -1.01 | 1.03 | 1.1 |
| pyrimidine nucleoside metabolic process | 10 | -1.32 | 1.14 | 1.04 | 1.22 | 1.27 | -1.25 | -1.09 |
| pyrimidine nucleotide metabolic process | 13 | 1.09 | 1.09 | 1.18 | 1.27 | 1.29 | -1.05 | -1.17 |
| 'de novo' pyrimidine base biosynthetic process | 6 | 1.66 | -1.0 | 1.02 | -1.03 | -1.03 | 1.11 | 1.08 |
| pyrimidine base catabolic process | 3 | 2.92 | -1.42 | -1.24 | -1.35 | -1.35 | 1.45 | 1.16 |
| uracil catabolic process | 1 | 1.44 | -1.11 | -1.15 | 1.16 | -1.17 | -1.1 | -1.2 |
| cytidine catabolic process | 3 | -2.8 | 1.04 | -1.39 | 1.2 | 1.32 | -1.87 | -1.06 |
| pyrimidine nucleotide biosynthetic process | 11 | 1.06 | 1.14 | 1.21 | 1.24 | 1.28 | -1.02 | -1.1 |
| UMP biosynthetic process | 3 | 1.1 | 1.3 | 1.05 | 1.25 | 1.1 | -1.22 | -1.06 |
| UTP biosynthetic process | 4 | 1.0 | 1.09 | 1.36 | 1.22 | 1.36 | 1.06 | -1.14 |
| TMP biosynthetic process | 1 | 1.02 | 1.09 | 1.32 | 1.32 | 1.3 | -1.29 | -1.13 |
| dTMP biosynthetic process | 2 | 1.05 | 1.03 | -1.1 | 1.26 | 1.31 | 1.33 | -1.09 |
| dTDP biosynthetic process | 1 | 1.27 | 1.13 | 1.91 | 1.21 | 1.41 | -1.17 | -1.08 |
| CTP biosynthetic process | 4 | 1.0 | 1.09 | 1.36 | 1.22 | 1.36 | 1.06 | -1.14 |
| DNA metabolic process | 257 | 1.17 | 1.09 | 1.23 | 1.13 | 1.3 | 1.15 | 1.06 |
| DNA-dependent DNA replication | 46 | 1.03 | 1.13 | 1.18 | 1.17 | 1.36 | 1.14 | -1.01 |
| mitochondrial DNA replication | 5 | -1.01 | 1.2 | 1.25 | 1.15 | 1.21 | 1.2 | -1.15 |
| DNA topological change | 7 | 1.09 | 1.2 | 1.22 | 1.21 | 1.33 | 1.09 | 1.23 |
| DNA ligation | 2 | 1.6 | 1.14 | 1.36 | 1.04 | 1.35 | 1.44 | 1.13 |
| pre-replicative complex assembly | 7 | -1.21 | 1.08 | 1.17 | 1.27 | 1.62 | -1.3 | -1.19 |
| DNA unwinding involved in replication | 2 | -1.01 | 1.27 | 1.01 | -1.0 | -1.05 | 1.15 | 1.27 |
| DNA replication, synthesis of RNA primer | 2 | 1.15 | 1.13 | 1.17 | 1.19 | 1.37 | 1.14 | -1.16 |
| DNA-dependent DNA replication initiation | 13 | -1.05 | 1.12 | 1.14 | 1.28 | 1.49 | -1.11 | -1.11 |
| DNA strand elongation involved in DNA replication | 3 | -1.21 | -1.08 | 1.4 | 1.14 | 1.61 | -1.08 | -1.25 |
| leading strand elongation | 1 | -1.9 | -1.01 | 1.54 | 1.17 | 1.67 | -2.38 | -1.41 |
| regulation of DNA replication | 17 | -1.04 | 1.13 | 1.23 | 1.15 | 1.36 | -1.13 | 1.15 |
| DNA amplification | 20 | 1.07 | 1.2 | 1.36 | 1.18 | 1.45 | 1.1 | 1.11 |
| RNA-dependent DNA replication | 1 | 1.54 | 1.82 | 1.92 | -1.04 | 1.87 | 1.15 | 2.39 |
| DNA repair | 93 | 1.23 | 1.08 | 1.27 | 1.13 | 1.31 | 1.19 | 1.06 |
| base-excision repair | 6 | 1.51 | -1.01 | 1.25 | 1.11 | 1.19 | 1.33 | -1.13 |
| base-excision repair, AP site formation | 1 | 1.23 | 1.13 | 1.14 | 1.14 | 1.15 | 1.01 | 1.07 |
| nucleotide-excision repair | 24 | 1.23 | 1.1 | 1.38 | 1.15 | 1.33 | 1.2 | 1.06 |
| nucleotide-excision repair, DNA incision, 5'-to lesion | 1 | 1.98 | 1.16 | 1.33 | 1.18 | 1.39 | 1.07 | 1.02 |
| mismatch repair | 8 | 1.32 | 1.07 | 1.23 | 1.15 | 1.41 | 1.18 | -1.02 |
| postreplication repair | 6 | 1.36 | 1.02 | 1.3 | 1.15 | 1.44 | 1.29 | 1.03 |
| double-strand break repair | 19 | 1.08 | 1.18 | 1.26 | 1.17 | 1.32 | -1.01 | 1.13 |
| double-strand break repair via nonhomologous end joining | 4 | -1.13 | 1.35 | 1.38 | 1.24 | 1.37 | -1.21 | 1.19 |
| DNA modification | 4 | 1.35 | 1.09 | 1.41 | 1.21 | 1.32 | 1.35 | 1.17 |
| DNA alkylation | 3 | 1.44 | 1.12 | 1.49 | 1.07 | 1.37 | 1.47 | 1.26 |
| DNA methylation | 3 | 1.44 | 1.12 | 1.49 | 1.07 | 1.37 | 1.47 | 1.26 |
| DNA dealkylation involved in DNA repair | 1 | 1.12 | -1.01 | 1.19 | 1.74 | 1.2 | 1.06 | -1.09 |
| DNA catabolic process | 9 | 1.71 | -1.1 | 1.05 | -1.12 | 1.18 | 1.29 | 1.41 |
| DNA fragmentation involved in apoptotic nuclear change | 3 | 1.86 | 1.09 | 1.48 | 1.14 | 1.43 | 1.6 | 1.67 |
| DNA recombination | 40 | 1.06 | 1.13 | 1.16 | 1.14 | 1.25 | 1.02 | 1.05 |
| transposition, DNA-mediated | 1 | 1.07 | -1.04 | -1.16 | 1.02 | 1.48 | -1.27 | -1.22 |
| DNA packaging | 70 | 1.1 | 1.09 | 1.27 | 1.13 | 1.3 | 1.1 | 1.1 |
| chromatin organization | 143 | 1.2 | 1.13 | 1.29 | 1.13 | 1.31 | 1.21 | 1.19 |
| DNA replication-independent nucleosome assembly | 1 | -1.49 | 1.47 | -1.04 | 1.39 | 1.42 | -1.65 | -1.06 |
| chromatin remodeling | 35 | 1.33 | 1.21 | 1.43 | 1.13 | 1.4 | 1.41 | 1.3 |
| chromatin silencing | 40 | 1.29 | 1.14 | 1.31 | 1.13 | 1.32 | 1.3 | 1.18 |
| chromatin-mediated maintenance of transcription | 8 | 1.92 | 1.19 | 1.53 | 1.04 | 1.43 | 1.72 | 1.45 |
| chromatin insulator sequence binding | 7 | 1.43 | 1.12 | 1.23 | 1.26 | 1.29 | 1.26 | 1.14 |
| establishment of chromatin silencing | 1 | 1.04 | -1.03 | 1.41 | 1.25 | 1.4 | 1.24 | -1.08 |
| chromatin silencing at telomere | 2 | 1.51 | 1.1 | 1.54 | 1.1 | 1.65 | 2.14 | 1.15 |
| regulation of gene expression by genetic imprinting | 1 | 1.01 | 1.1 | 1.09 | 1.14 | -1.01 | 1.17 | 1.11 |
| transcription, DNA-dependent | 149 | 1.27 | 1.12 | 1.22 | 1.16 | 1.25 | 1.16 | 1.13 |
| transcription initiation, DNA-dependent | 68 | 1.28 | 1.1 | 1.25 | 1.16 | 1.25 | 1.15 | 1.14 |
| transcription elongation, DNA-dependent | 7 | 1.13 | 1.24 | 1.33 | 1.14 | 1.27 | 1.28 | 1.28 |
| regulation of transcription, DNA-dependent | 671 | 1.13 | 1.08 | 1.17 | 1.06 | 1.16 | 1.15 | 1.13 |
| regulation of transcription from RNA polymerase I promoter | 1 | 1.03 | 1.42 | 1.12 | 1.32 | 1.22 | 1.06 | 1.31 |
| regulation of transcription from RNA polymerase II promoter | 306 | 1.09 | 1.06 | 1.16 | 1.04 | 1.13 | 1.13 | 1.11 |
| regulation of transcription from RNA polymerase III promoter | 5 | 1.56 | 1.19 | 1.46 | 1.24 | 1.48 | 1.38 | 1.12 |
| rRNA processing | 27 | 1.21 | 1.16 | 1.21 | 1.32 | 1.31 | 1.02 | 1.04 |
| transcription initiation from RNA polymerase II promoter | 62 | 1.34 | 1.12 | 1.27 | 1.18 | 1.28 | 1.18 | 1.11 |
| transcription elongation from RNA polymerase II promoter | 6 | 1.12 | 1.24 | 1.29 | 1.14 | 1.22 | 1.21 | 1.27 |
| mRNA capping | 2 | 1.43 | 1.2 | 1.32 | 1.24 | 1.14 | 1.18 | 1.16 |
| mRNA splice site selection | 8 | 1.08 | 1.11 | 1.22 | 1.24 | 1.25 | 1.14 | -1.09 |
| mRNA polyadenylation | 16 | 1.4 | 1.12 | 1.26 | 1.13 | 1.32 | 1.72 | 1.2 |
| mRNA cleavage | 15 | 1.21 | 1.25 | 1.41 | 1.3 | 1.43 | 1.28 | 1.15 |
| poly(A) RNA binding | 5 | 1.32 | 1.19 | 1.41 | 1.19 | 1.38 | 1.34 | 1.35 |
| adenosine to inosine editing | 1 | 1.13 | 1.14 | 1.55 | 1.01 | 1.4 | 2.0 | 1.26 |
| transcription initiation from RNA polymerase III promoter | 1 | 1.15 | 1.32 | 1.1 | 1.22 | 1.43 | 1.06 | 1.02 |
| tRNA splicing, via endonucleolytic cleavage and ligation | 3 | 1.93 | 1.15 | 1.42 | 1.2 | 1.36 | 1.44 | 1.34 |
| transcription from mitochondrial promoter | 3 | 1.29 | 1.23 | 1.41 | 1.12 | 1.47 | 1.73 | -1.1 |
| termination of mitochondrial transcription | 1 | 1.24 | 1.11 | 1.4 | 1.15 | 1.32 | 1.42 | -1.37 |
| RNA processing | 315 | 1.22 | 1.13 | 1.23 | 1.22 | 1.29 | 1.19 | 1.03 |
| mRNA processing | 228 | 1.21 | 1.12 | 1.23 | 1.22 | 1.31 | 1.2 | 1.04 |
| histone mRNA 3'-end processing | 8 | 1.38 | 1.09 | 1.19 | 1.27 | 1.38 | 1.29 | 1.05 |
| tRNA metabolic process | 74 | 1.1 | 1.19 | 1.2 | 1.18 | 1.17 | 1.07 | 1.01 |
| tRNA modification | 16 | 1.24 | 1.15 | 1.11 | 1.21 | 1.19 | 1.09 | -1.07 |
| RNA catabolic process | 30 | 1.13 | 1.15 | 1.32 | 1.17 | 1.34 | 1.34 | 1.22 |
| mRNA catabolic process | 24 | 1.1 | 1.16 | 1.33 | 1.18 | 1.37 | 1.31 | 1.2 |
| RNA localization | 99 | 1.01 | 1.16 | 1.28 | 1.14 | 1.27 | 1.16 | 1.09 |
| RNA import into nucleus | 1 | 1.13 | 1.03 | 1.18 | 1.15 | 1.1 | 1.24 | -1.22 |
| RNA export from nucleus | 20 | 1.14 | 1.1 | 1.18 | 1.19 | 1.21 | 1.08 | -1.04 |
| mRNA export from nucleus | 18 | 1.1 | 1.09 | 1.18 | 1.2 | 1.21 | 1.06 | -1.05 |
| snRNA export from nucleus | 2 | 1.9 | 1.18 | 1.33 | 1.29 | 1.62 | 1.23 | 1.13 |
| tRNA export from nucleus | 1 | 1.48 | 1.1 | 1.33 | 1.32 | 1.54 | -1.18 | -1.04 |
| translation | 279 | 1.2 | 1.1 | 1.19 | 1.11 | 1.18 | 1.26 | -1.04 |
| translational initiation | 52 | 1.32 | 1.17 | 1.24 | 1.09 | 1.16 | 1.33 | 1.15 |
| translational elongation | 18 | 1.46 | 1.07 | 1.16 | 1.06 | 1.21 | 1.3 | 1.01 |
| translational termination | 6 | 1.2 | 1.18 | 1.21 | 1.16 | 1.16 | 1.51 | 1.06 |
| regulation of translation | 53 | 1.42 | 1.08 | 1.27 | 1.03 | 1.3 | 1.38 | 1.28 |
| tRNA aminoacylation for protein translation | 41 | -1.07 | 1.25 | 1.27 | 1.14 | 1.19 | 1.05 | 1.09 |
| alanyl-tRNA aminoacylation | 3 | -1.05 | 1.37 | 1.07 | 1.17 | 1.09 | 1.18 | 1.07 |
| arginyl-tRNA aminoacylation | 3 | 1.35 | 1.15 | 1.32 | 1.15 | 1.24 | 1.15 | 1.05 |
| asparaginyl-tRNA aminoacylation | 2 | -1.04 | 1.15 | 1.12 | 1.09 | 1.19 | -1.05 | 1.03 |
| aspartyl-tRNA aminoacylation | 1 | -1.97 | 1.61 | 1.28 | -1.18 | 1.45 | -1.88 | 1.38 |
| cysteinyl-tRNA aminoacylation | 2 | -1.01 | 1.37 | 1.43 | 1.08 | 1.22 | 1.24 | 1.76 |
| glutamyl-tRNA aminoacylation | 2 | 1.05 | 1.23 | 1.54 | -1.01 | 1.6 | 1.42 | 1.27 |
| glutaminyl-tRNA aminoacylation | 2 | 1.01 | 1.2 | 1.18 | 1.26 | 1.25 | 1.22 | -1.02 |
| glycyl-tRNA aminoacylation | 3 | 1.01 | 1.17 | 1.37 | 1.21 | 1.21 | 1.27 | -1.07 |
| histidyl-tRNA aminoacylation | 1 | -1.32 | 1.37 | -1.07 | 1.31 | -1.14 | -1.74 | -1.19 |
| isoleucyl-tRNA aminoacylation | 2 | 1.35 | 1.22 | 1.54 | 1.02 | 1.38 | 1.61 | 1.38 |
| leucyl-tRNA aminoacylation | 2 | -1.17 | 1.4 | 1.1 | 1.12 | 1.07 | 1.1 | 1.24 |
| lysyl-tRNA aminoacylation | 1 | 1.12 | 1.29 | 1.39 | 1.11 | 1.29 | 1.02 | 1.14 |
| methionyl-tRNA aminoacylation | 2 | -1.59 | 1.18 | 1.28 | 1.38 | 1.17 | -1.27 | -1.03 |
| phenylalanyl-tRNA aminoacylation | 3 | -1.16 | 1.27 | 1.38 | 1.24 | 1.13 | -1.0 | -1.14 |
| prolyl-tRNA aminoacylation | 2 | -1.08 | 1.19 | 1.31 | 1.06 | 1.32 | 1.47 | 1.15 |
| seryl-tRNA aminoacylation | 3 | -1.3 | 1.34 | 1.25 | 1.17 | 1.28 | -1.22 | -1.0 |
| threonyl-tRNA aminoacylation | 2 | -1.65 | 1.24 | 1.06 | 1.13 | -1.08 | -1.17 | 1.02 |
| tryptophanyl-tRNA aminoacylation | 2 | 1.09 | 1.09 | 1.24 | 1.11 | 1.12 | 1.12 | 1.03 |
| tyrosyl-tRNA aminoacylation | 2 | 1.14 | 1.27 | 1.29 | 1.14 | 1.19 | 1.03 | -1.04 |
| valyl-tRNA aminoacylation | 2 | -1.62 | 1.31 | 1.29 | 1.15 | 1.04 | -1.25 | 1.17 |
| regulation of translational initiation | 10 | 1.76 | -1.0 | 1.08 | -1.04 | 1.23 | 1.53 | 1.27 |
| regulation of translational initiation by iron | 2 | 1.36 | 1.24 | 1.32 | -1.06 | 1.1 | 1.69 | 1.16 |
| regulation of translational elongation | 3 | 1.5 | 1.19 | 1.07 | 1.08 | 1.04 | 1.01 | 1.17 |
| regulation of translational termination | 1 | 1.71 | 1.01 | 1.01 | -1.13 | 1.21 | 1.32 | 1.15 |
| translational readthrough | 2 | 1.54 | 1.17 | 1.08 | 1.41 | 1.45 | -1.1 | -1.08 |
| translational frameshifting | 1 | 1.71 | 1.01 | 1.01 | -1.13 | 1.21 | 1.32 | 1.15 |
| 'de novo' protein folding | 13 | 1.11 | 1.05 | 1.16 | 1.13 | 1.14 | -1.02 | -1.03 |
| protein complex assembly | 80 | 1.0 | 1.09 | 1.26 | 1.1 | 1.24 | 1.12 | -1.01 |
| protein modification process | 623 | 1.16 | 1.1 | 1.22 | 1.07 | 1.22 | 1.25 | 1.13 |
| signal peptide processing | 3 | 1.12 | -1.0 | 1.0 | -1.02 | -1.02 | -1.01 | 1.75 |
| protein phosphorylation | 241 | 1.17 | 1.1 | 1.26 | 1.06 | 1.28 | 1.37 | 1.18 |
| negative regulation of protein kinase activity | 12 | 1.53 | 1.14 | 1.46 | 1.09 | 1.41 | 1.69 | 1.32 |
| protein dephosphorylation | 81 | 1.24 | 1.1 | 1.23 | 1.06 | 1.22 | 1.31 | 1.13 |
| protein ADP-ribosylation | 10 | 1.1 | 1.07 | 1.16 | 1.06 | 1.17 | -1.04 | 1.01 |
| protein acetylation | 16 | 1.31 | 1.25 | 1.45 | 1.18 | 1.34 | 1.36 | 1.25 |
| N-terminal protein amino acid acetylation | 1 | -1.5 | 1.47 | 2.65 | 1.15 | 1.31 | -1.14 | -1.06 |
| internal protein amino acid acetylation | 14 | 1.4 | 1.24 | 1.42 | 1.18 | 1.37 | 1.44 | 1.29 |
| protein deacetylation | 9 | 1.34 | 1.15 | 1.08 | 1.23 | 1.2 | 1.41 | 1.12 |
| protein methylation | 32 | 1.29 | 1.19 | 1.36 | 1.15 | 1.36 | 1.34 | 1.2 |
| C-terminal protein methylation | 1 | -2.13 | 1.18 | 1.09 | 1.41 | 1.8 | -1.71 | -1.76 |
| protein demethylation | 7 | 1.18 | 1.25 | 1.72 | 1.13 | 1.59 | 1.68 | 1.3 |
| protein glycosylation | 51 | 1.14 | 1.04 | 1.06 | 1.04 | 1.1 | 1.12 | 1.22 |
| protein N-linked glycosylation | 14 | 1.33 | 1.02 | 1.11 | 1.03 | 1.13 | 1.55 | 1.26 |
| dolichol-linked oligosaccharide biosynthetic process | 2 | 1.08 | 1.14 | 1.16 | -1.01 | 1.12 | 1.06 | 1.17 |
| oligosaccharide-lipid intermediate biosynthetic process | 1 | -1.04 | -1.0 | 1.06 | -1.08 | -1.02 | -1.02 | -1.17 |
| N-glycan processing | 1 | 3.13 | 1.28 | 1.73 | 1.05 | 1.33 | 2.12 | 2.37 |
| protein O-linked glycosylation | 10 | -1.06 | 1.11 | 1.08 | 1.09 | 1.19 | -1.37 | 1.29 |
| protein lipidation | 50 | -1.11 | 1.0 | 1.04 | 1.04 | 1.13 | -1.02 | -1.14 |
| N-terminal protein lipidation | 2 | -1.18 | 1.31 | 1.73 | 1.36 | 1.77 | -1.01 | -1.01 |
| N-terminal protein myristoylation | 1 | -1.49 | 1.29 | 1.71 | 1.24 | 1.7 | 1.05 | 1.0 |
| N-terminal protein palmitoylation | 1 | 1.07 | 1.32 | 1.75 | 1.48 | 1.85 | -1.07 | -1.03 |
| protein prenylation | 4 | -1.32 | 1.12 | 1.23 | 1.09 | 1.23 | -1.12 | 1.41 |
| protein geranylgeranylation | 1 | 1.07 | 1.22 | 1.65 | 1.13 | 1.53 | -1.02 | -1.05 |
| GPI anchor metabolic process | 25 | -1.3 | -1.04 | 1.04 | -1.0 | 1.2 | 1.0 | -1.25 |
| GPI anchor biosynthetic process | 22 | -1.36 | -1.06 | -1.0 | -1.0 | 1.15 | -1.03 | -1.34 |
| GPI anchor release | 1 | 1.09 | 1.12 | 1.02 | 1.1 | 1.11 | 1.04 | 1.13 |
| proteolysis | 624 | -1.68 | -1.23 | -1.28 | -1.03 | -1.1 | -1.65 | -1.4 |
| membrane protein ectodomain proteolysis | 2 | -1.1 | 1.22 | 1.43 | 1.31 | 1.8 | -1.31 | 1.17 |
| protein processing | 33 | 1.26 | -1.01 | 1.14 | -1.0 | 1.11 | 1.16 | 1.21 |
| proteolysis involved in cellular protein catabolic process | 106 | 1.15 | 1.12 | 1.31 | 1.13 | 1.28 | 1.07 | 1.08 |
| ubiquitin-dependent protein catabolic process | 92 | 1.16 | 1.13 | 1.33 | 1.13 | 1.29 | 1.1 | 1.13 |
| protein monoubiquitination | 1 | -1.01 | -1.28 | 2.49 | -1.24 | 1.59 | -1.44 | -1.52 |
| misfolded or incompletely synthesized protein catabolic process | 1 | 2.66 | -1.19 | -1.04 | 1.2 | -1.17 | 1.37 | 1.41 |
| glycoprotein catabolic process | 1 | -1.09 | 1.27 | 1.39 | 1.28 | 1.54 | -1.1 | 1.18 |
| protein deglycosylation | 1 | 3.0 | -1.25 | -1.04 | 1.43 | 1.17 | 2.32 | 1.34 |
| peptide metabolic process | 9 | -1.21 | 1.04 | -1.26 | 1.07 | 1.11 | -1.18 | 1.4 |
| cellular amino acid metabolic process | 161 | -1.02 | 1.04 | -1.09 | -1.01 | 1.01 | 1.14 | -1.04 |
| alanine metabolic process | 1 | 1.7 | 1.2 | 1.13 | 1.08 | -1.16 | -1.08 | 1.29 |
| alanine biosynthetic process | 1 | 1.7 | 1.2 | 1.13 | 1.08 | -1.16 | -1.08 | 1.29 |
| arginine metabolic process | 5 | -5.16 | 1.27 | -5.44 | 1.2 | 1.07 | -3.1 | -1.21 |
| arginine biosynthetic process | 2 | -22.15 | 1.64 | -12.84 | 1.4 | 1.07 | -7.77 | -1.97 |
| arginine catabolic process | 2 | -3.2 | -1.06 | -5.13 | -1.12 | -1.07 | -1.58 | 1.05 |
| asparagine metabolic process | 2 | -1.07 | 1.2 | 1.14 | 1.26 | 1.17 | 1.27 | -1.14 |
| asparagine biosynthetic process | 2 | -1.07 | 1.2 | 1.14 | 1.26 | 1.17 | 1.27 | -1.14 |
| aspartate metabolic process | 2 | 1.83 | 1.18 | -1.32 | -1.0 | 1.19 | 1.57 | 1.14 |
| cysteine metabolic process | 3 | 1.38 | 1.11 | 1.22 | -1.23 | -2.05 | -1.02 | 1.55 |
| cysteine biosynthetic process from serine | 1 | 1.25 | 1.1 | 1.48 | -1.98 | -7.11 | -1.17 | 2.59 |
| glutamate metabolic process | 10 | -1.06 | -1.22 | -1.57 | -1.37 | 1.12 | 1.73 | -1.05 |
| glutamate biosynthetic process | 5 | 1.59 | -1.31 | -1.43 | -1.5 | -1.34 | 2.96 | 1.23 |
| glutamate catabolic process | 4 | -1.09 | -1.24 | 1.09 | -1.34 | 1.82 | 1.75 | -1.68 |
| glutamine metabolic process | 5 | -1.72 | -1.12 | -1.2 | -1.13 | 1.54 | -1.57 | -3.03 |
| glutamine biosynthetic process | 2 | 1.91 | -1.99 | 2.01 | -1.44 | 1.52 | -1.73 | -1.25 |
| glutamine catabolic process | 1 | -1.12 | 1.14 | 1.63 | 1.21 | 1.33 | 1.0 | 1.05 |
| glycine metabolic process | 9 | 1.41 | 1.01 | 1.25 | -1.01 | -1.01 | 1.77 | -1.06 |
| glycine biosynthetic process | 1 | 2.25 | -1.1 | -1.03 | 1.28 | 1.09 | 2.14 | -1.24 |
| glycine catabolic process | 6 | 1.21 | 1.07 | 1.39 | -1.04 | -1.02 | 1.86 | -1.02 |
| isoleucine metabolic process | 1 | 1.48 | 1.28 | 1.32 | 1.05 | 1.07 | 1.16 | -1.01 |
| isoleucine catabolic process | 1 | 1.48 | 1.28 | 1.32 | 1.05 | 1.07 | 1.16 | -1.01 |
| leucine metabolic process | 4 | 1.15 | 1.54 | 1.31 | 1.24 | 1.31 | 1.45 | 1.1 |
| leucine catabolic process | 1 | 1.14 | 1.39 | 1.15 | 1.11 | 1.04 | 1.05 | 1.01 |
| lysine metabolic process | 1 | -1.51 | -1.6 | -1.42 | 1.8 | -1.74 | 2.0 | -2.55 |
| lysine catabolic process | 1 | -1.51 | -1.6 | -1.42 | 1.8 | -1.74 | 2.0 | -2.55 |
| methionine metabolic process | 6 | 1.26 | -1.05 | -1.1 | -1.03 | -1.11 | 1.66 | -1.23 |
| S-adenosylmethionine biosynthetic process | 1 | 1.6 | 1.05 | 1.23 | -1.23 | 1.41 | 1.78 | 1.98 |
| L-phenylalanine metabolic process | 4 | 1.52 | -1.14 | -1.01 | -1.01 | -1.02 | 2.14 | -1.13 |
| L-phenylalanine catabolic process | 4 | 1.52 | -1.14 | -1.01 | -1.01 | -1.02 | 2.14 | -1.13 |
| proline metabolic process | 7 | -1.43 | -1.33 | -1.79 | -1.62 | -1.41 | 1.63 | 1.05 |
| proline biosynthetic process | 4 | -1.95 | -1.03 | -1.89 | -1.37 | -1.23 | 1.12 | 1.21 |
| proline catabolic process | 2 | 1.19 | -2.55 | -2.04 | -2.77 | -1.87 | 5.01 | -1.07 |
| L-serine metabolic process | 5 | 2.33 | 1.11 | -1.22 | 1.0 | -1.34 | 1.61 | 1.43 |
| L-serine biosynthetic process | 4 | 2.73 | 1.12 | -1.41 | 1.19 | 1.13 | 1.89 | 1.23 |
| threonine metabolic process | 1 | 1.48 | 1.28 | 1.32 | 1.05 | 1.07 | 1.16 | -1.01 |
| threonine catabolic process | 1 | 1.48 | 1.28 | 1.32 | 1.05 | 1.07 | 1.16 | -1.01 |
| tryptophan metabolic process | 7 | 1.13 | -1.34 | -1.47 | -1.15 | -1.37 | 1.23 | 1.22 |
| tryptophan catabolic process | 2 | 1.18 | -1.21 | -1.17 | -1.24 | -1.21 | 1.62 | -1.18 |
| tyrosine metabolic process | 8 | 1.53 | -1.29 | -1.23 | -1.14 | -1.34 | 1.56 | 1.15 |
| tyrosine catabolic process | 3 | 1.31 | -1.03 | 1.15 | 1.15 | 1.06 | 1.47 | -1.07 |
| valine metabolic process | 3 | -1.09 | 1.41 | -1.02 | -1.03 | -1.14 | 1.15 | 1.23 |
| valine catabolic process | 1 | 1.48 | 1.28 | 1.32 | 1.05 | 1.07 | 1.16 | -1.01 |
| cellular modified amino acid metabolic process | 27 | -1.23 | 1.06 | -1.31 | 1.01 | -1.04 | -1.11 | -1.06 |
| cellular biogenic amine metabolic process | 29 | 1.1 | -1.1 | -1.15 | -1.04 | -1.12 | 1.08 | 1.25 |
| betaine metabolic process | 2 | -1.01 | -1.08 | -1.06 | -1.12 | -1.16 | 1.15 | -1.15 |
| betaine biosynthetic process | 1 | 1.3 | -1.08 | -1.07 | -1.13 | -1.19 | 1.96 | -1.06 |
| ethanolamine metabolic process | 1 | 1.07 | -1.05 | 2.34 | 1.23 | 1.45 | 3.99 | 1.1 |
| acetylcholine catabolic process | 1 | 2.27 | 1.23 | -5.17 | -1.59 | -2.2 | -9.9 | 9.42 |
| melanin metabolic process | 16 | 2.27 | -1.2 | -1.16 | -1.18 | 1.1 | 1.65 | 1.32 |
| melanin biosynthetic process from tyrosine | 4 | 1.82 | -1.08 | -1.09 | -1.03 | -1.18 | 1.82 | -1.13 |
| catecholamine metabolic process | 9 | -1.02 | -1.18 | -1.26 | -1.16 | -1.28 | -1.1 | 1.52 |
| dopamine biosynthetic process from tyrosine | 1 | 1.23 | -4.96 | -5.74 | -3.75 | -6.41 | -1.0 | 6.11 |
| indolalkylamine metabolic process | 8 | -1.18 | -1.2 | -1.54 | -1.13 | -1.45 | -1.17 | 1.22 |
| serotonin biosynthetic process from tryptophan | 2 | 1.67 | -2.04 | -2.31 | -1.94 | -2.17 | 1.48 | 3.91 |
| activation of tryptophan 5-monooxygenase activity | 1 | 2.28 | 1.19 | 1.07 | -1.0 | 1.37 | 2.2 | 2.5 |
| octopamine biosynthetic process | 1 | 1.3 | 1.0 | -1.05 | -1.09 | 1.04 | -1.06 | -1.0 |
| ornithine metabolic process | 3 | -2.2 | 1.14 | -2.13 | 1.11 | -1.02 | -1.24 | -1.28 |
| polyamine metabolic process | 6 | -1.02 | -1.06 | -1.06 | 1.08 | -1.03 | 1.44 | -1.28 |
| polyamine biosynthetic process | 5 | 1.03 | -1.1 | -1.08 | 1.04 | -1.04 | 1.59 | -1.28 |
| spermine biosynthetic process | 2 | 1.12 | 1.21 | 1.31 | 1.22 | 1.2 | 1.92 | 1.01 |
| polyamine catabolic process | 1 | -1.3 | 1.12 | 1.08 | 1.3 | 1.03 | -1.14 | -1.27 |
| protein targeting | 109 | 1.05 | 1.18 | 1.26 | 1.14 | 1.21 | 1.12 | 1.14 |
| NLS-bearing substrate import into nucleus | 7 | -1.09 | 1.31 | 1.37 | 1.28 | 1.56 | 1.0 | 1.17 |
| protein export from nucleus | 5 | 1.19 | 1.11 | 2.28 | 1.15 | 1.72 | 1.71 | 1.32 |
| protein targeting to membrane | 25 | -1.09 | 1.24 | 1.08 | 1.08 | -1.04 | -1.21 | 1.13 |
| cotranslational protein targeting to membrane | 16 | -1.27 | 1.37 | -1.06 | 1.05 | -1.21 | -1.6 | 1.25 |
| SRP-dependent cotranslational protein targeting to membrane | 15 | -1.21 | 1.37 | -1.05 | 1.04 | -1.19 | -1.54 | 1.26 |
| SRP-dependent cotranslational protein targeting to membrane, translocation | 5 | -1.62 | 1.28 | -1.17 | -1.01 | -1.28 | -1.9 | 1.13 |
| SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition | 1 | -1.09 | 1.62 | 1.41 | 1.12 | 1.35 | 1.19 | 1.81 |
| protein retention in ER lumen | 3 | -1.9 | 1.28 | -1.09 | 1.08 | -1.19 | -2.08 | 1.34 |
| protein targeting to lysosome | 3 | 1.19 | 1.18 | 1.82 | 1.21 | 1.4 | 1.29 | 1.24 |
| protein targeting to vacuole | 4 | 1.47 | 1.26 | 2.02 | 1.17 | 1.52 | 1.6 | 1.47 |
| protein targeting to mitochondrion | 28 | 1.07 | 1.09 | 1.21 | 1.13 | 1.12 | 1.18 | -1.1 |
| protein processing involved in protein targeting to mitochondrion | 2 | 1.16 | 1.04 | 1.32 | 1.16 | 1.03 | 1.09 | -1.27 |
| lipid metabolic process | 302 | -1.17 | -1.16 | -1.23 | -1.08 | -1.04 | -1.3 | -1.11 |
| lipid binding | 177 | 1.05 | -1.11 | 1.02 | -1.03 | 1.14 | 1.13 | 1.06 |
| fatty acid metabolic process | 49 | 1.06 | -1.23 | -1.39 | -1.08 | -1.07 | -1.23 | -1.05 |
| fatty acid biosynthetic process | 25 | 1.1 | -1.38 | -1.52 | -1.26 | -1.29 | 1.04 | 1.14 |
| fatty acid beta-oxidation | 12 | -1.2 | 1.1 | -1.58 | 1.06 | 1.17 | -2.29 | -1.36 |
| unsaturated fatty acid biosynthetic process | 1 | -1.74 | 1.11 | -2.01 | 1.51 | 1.27 | -1.87 | -1.55 |
| acyl-CoA metabolic process | 6 | 2.01 | 1.03 | 1.27 | 1.12 | 1.42 | 1.33 | 1.06 |
| neutral lipid metabolic process | 10 | 1.74 | -1.01 | 1.02 | -1.04 | 1.58 | -1.11 | -1.1 |
| acylglycerol metabolic process | 10 | 1.74 | -1.01 | 1.02 | -1.04 | 1.58 | -1.11 | -1.1 |
| triglyceride metabolic process | 7 | 1.7 | 1.03 | -1.06 | -1.02 | 1.5 | 1.09 | -1.06 |
| triglyceride mobilization | 1 | -2.11 | 1.69 | -1.72 | -1.16 | 2.47 | -2.75 | -1.82 |
| membrane lipid metabolic process | 28 | -2.18 | -1.19 | -1.19 | 1.18 | 1.02 | -2.06 | -1.41 |
| phospholipid metabolic process | 93 | 1.02 | -1.05 | 1.02 | 1.02 | 1.23 | 1.02 | -1.04 |
| phosphatidylethanolamine biosynthetic process | 1 | 1.07 | -1.05 | 2.34 | 1.23 | 1.45 | 3.99 | 1.1 |
| glycerophospholipid metabolic process | 66 | 1.09 | 1.03 | 1.13 | 1.01 | 1.25 | 1.18 | 1.06 |
| diacylglycerol biosynthetic process | 2 | 4.42 | -1.06 | 2.12 | -1.4 | 1.56 | 1.46 | 1.74 |
| phosphatidylglycerol biosynthetic process | 1 | -3.36 | 1.4 | 1.32 | 1.25 | 1.35 | -2.25 | 1.21 |
| phosphatidylcholine biosynthetic process | 1 | -1.0 | -1.38 | 1.09 | 1.05 | 1.26 | -1.4 | 2.06 |
| phosphatidylserine metabolic process | 1 | 1.61 | 1.37 | 2.22 | 1.0 | 1.13 | 1.67 | 1.83 |
| phosphatidylserine biosynthetic process | 1 | 1.61 | 1.37 | 2.22 | 1.0 | 1.13 | 1.67 | 1.83 |
| phosphatidylinositol biosynthetic process | 24 | -1.24 | -1.05 | 1.03 | -1.01 | 1.12 | -1.0 | -1.25 |
| glycerol ether metabolic process | 26 | 1.41 | -1.13 | -1.09 | -1.07 | 1.17 | -1.17 | 1.01 |
| glycolipid metabolic process | 8 | 1.02 | 1.23 | 1.09 | 1.02 | 1.05 | -1.13 | 1.74 |
| sphingolipid metabolic process | 24 | -2.32 | -1.33 | -1.28 | 1.19 | 1.03 | -2.16 | -1.6 |
| ceramide metabolic process | 7 | -1.82 | -1.06 | 1.22 | 1.15 | 1.12 | -2.11 | -1.7 |
| glycosylceramide metabolic process | 1 | -1.02 | 1.14 | 1.14 | 1.39 | 1.15 | 1.04 | -1.06 |
| glucosylceramide metabolic process | 1 | -1.02 | 1.14 | 1.14 | 1.39 | 1.15 | 1.04 | -1.06 |
| glucosylceramide catabolic process | 1 | -1.02 | 1.14 | 1.14 | 1.39 | 1.15 | 1.04 | -1.06 |
| sphingomyelin metabolic process | 5 | -5.26 | -3.33 | -4.18 | 1.39 | -1.08 | -3.6 | -4.11 |
| sphingomyelin catabolic process | 3 | -14.19 | -8.88 | -14.23 | 1.69 | -1.38 | -9.59 | -10.93 |
| sphingomyelin biosynthetic process | 1 | -1.01 | 1.47 | 1.74 | -1.23 | 1.22 | 1.11 | 1.04 |
| glycosphingolipid metabolic process | 5 | 1.35 | 1.03 | 1.0 | 1.01 | 1.04 | 1.17 | 1.95 |
| glycosphingolipid biosynthetic process | 4 | 1.47 | 1.01 | -1.03 | -1.08 | 1.02 | 1.2 | 2.34 |
| icosanoid metabolic process | 1 | -1.74 | 1.11 | -2.01 | 1.51 | 1.27 | -1.87 | -1.55 |
| leukotriene metabolic process | 1 | -1.74 | 1.11 | -2.01 | 1.51 | 1.27 | -1.87 | -1.55 |
| steroid biosynthetic process | 18 | 1.4 | -1.12 | -1.0 | -1.04 | 1.07 | 1.48 | 1.09 |
| cholesterol biosynthetic process | 1 | 2.16 | -1.1 | 1.03 | -1.13 | 1.06 | 2.2 | 1.18 |
| ecdysone biosynthetic process | 7 | 1.01 | 1.04 | 1.04 | 1.09 | -1.04 | 1.04 | 1.01 |
| steroid catabolic process | 2 | -1.06 | 1.06 | 1.18 | 1.13 | 1.19 | 1.01 | -1.1 |
| cholesterol catabolic process | 1 | -1.19 | 1.24 | 1.2 | 1.43 | 1.53 | -1.12 | -1.13 |
| sesquiterpenoid metabolic process | 6 | -1.16 | -1.26 | -1.56 | -1.5 | -1.43 | -2.35 | 1.61 |
| juvenile hormone metabolic process | 6 | -1.16 | -1.26 | -1.56 | -1.5 | -1.43 | -2.35 | 1.61 |
| juvenile hormone biosynthetic process | 2 | 1.77 | -2.92 | -3.1 | -3.09 | -1.69 | -1.43 | 2.62 |
| juvenile hormone catabolic process | 4 | -1.67 | 1.21 | -1.1 | -1.05 | -1.31 | -3.02 | 1.26 |
| isoprenoid metabolic process | 19 | 1.13 | -1.14 | -1.16 | -1.06 | 1.01 | -1.18 | 1.11 |
| terpenoid metabolic process | 11 | -1.32 | -1.26 | -1.61 | -1.13 | -1.21 | -1.86 | 1.01 |
| cuticle hydrocarbon biosynthetic process | 3 | 1.33 | -1.03 | -1.15 | 1.2 | 1.17 | 1.22 | 1.02 |
| cellular aromatic compound metabolic process | 72 | 1.45 | -1.01 | -1.02 | -1.0 | -1.03 | 1.22 | 1.11 |
| eye pigment biosynthetic process | 29 | 1.05 | -1.06 | -1.17 | 1.17 | 1.04 | -1.09 | -1.16 |
| ommochrome biosynthetic process | 18 | 1.35 | 1.13 | 1.12 | 1.06 | 1.06 | 1.08 | 1.16 |
| pteridine biosynthetic process | 5 | 2.4 | -1.22 | -1.26 | -1.24 | -1.16 | -1.04 | 1.7 |
| tetrahydrobiopterin biosynthetic process | 4 | 7.84 | -1.48 | -1.41 | -1.58 | -1.23 | 1.76 | 2.48 |
| one-carbon metabolic process | 15 | -1.73 | -1.28 | 1.21 | -1.29 | 1.06 | 1.32 | -1.1 |
| coenzyme metabolic process | 86 | 1.32 | 1.09 | 1.09 | 1.0 | 1.09 | 1.34 | 1.06 |
| prosthetic group metabolic process | 5 | 1.14 | 1.31 | 1.37 | 1.14 | 1.16 | -1.34 | 1.2 |
| oxidoreduction coenzyme metabolic process | 16 | 1.6 | 1.17 | -1.26 | 1.03 | 1.09 | 1.19 | 1.2 |
| NADH metabolic process | 1 | -1.32 | -1.9 | 1.3 | -2.14 | 3.35 | 4.87 | -4.48 |
| NADP metabolic process | 7 | 2.13 | 1.17 | -1.85 | 1.03 | -1.09 | 1.03 | 1.53 |
| NADPH regeneration | 7 | 2.13 | 1.17 | -1.85 | 1.03 | -1.09 | 1.03 | 1.53 |
| ubiquinone metabolic process | 5 | 1.12 | 1.18 | 1.13 | 1.21 | 1.09 | 1.08 | -1.08 |
| ubiquinone biosynthetic process | 4 | 1.15 | 1.16 | 1.14 | 1.19 | 1.12 | 1.3 | -1.07 |
| lipoamide metabolic process | 1 | -1.4 | -1.14 | 2.27 | -1.25 | -1.48 | 1.94 | -1.5 |
| glutathione biosynthetic process | 3 | -2.99 | 1.44 | -1.36 | 1.22 | -1.32 | -2.2 | 1.15 |
| nucleoside phosphate metabolic process | 167 | 1.11 | 1.05 | 1.05 | 1.02 | 1.07 | 1.16 | 1.0 |
| ATP biosynthetic process | 25 | 1.34 | 1.04 | 1.24 | -1.05 | 1.09 | 1.66 | -1.1 |
| folic acid-containing compound metabolic process | 8 | 1.68 | -1.27 | -1.29 | -1.11 | -1.33 | 1.27 | 1.0 |
| vitamin metabolic process | 11 | 1.53 | 1.22 | -1.23 | -1.04 | -1.18 | 1.35 | 1.13 |
| water-soluble vitamin metabolic process | 7 | 1.81 | 1.34 | -1.5 | -1.06 | -1.3 | 1.43 | 1.2 |
| riboflavin metabolic process | 2 | 1.78 | 2.05 | -2.11 | -2.16 | -2.52 | 2.14 | 1.44 |
| thiamine metabolic process | 1 | 1.06 | 1.62 | -1.26 | 1.22 | -1.14 | -1.11 | 1.12 |
| fat-soluble vitamin metabolic process | 3 | 1.15 | 1.02 | 1.22 | -1.01 | 1.05 | 1.32 | 1.08 |
| vitamin A metabolic process | 2 | 1.02 | 1.01 | 1.08 | 1.02 | -1.08 | 1.12 | -1.06 |
| Mo-molybdopterin cofactor biosynthetic process | 5 | 1.14 | 1.31 | 1.37 | 1.14 | 1.16 | -1.34 | 1.2 |
| porphyrin-containing compound metabolic process | 9 | 1.28 | 1.21 | 1.24 | 1.17 | 1.12 | 1.19 | -1.04 |
| porphyrin-containing compound biosynthetic process | 8 | 1.22 | 1.23 | 1.16 | 1.18 | 1.09 | 1.1 | -1.06 |
| protoporphyrinogen IX biosynthetic process | 1 | 1.62 | 1.26 | 1.09 | -1.07 | 1.01 | 1.21 | 1.32 |
| heme biosynthetic process | 8 | 1.22 | 1.23 | 1.16 | 1.18 | 1.09 | 1.1 | -1.06 |
| heme a biosynthetic process | 1 | -1.08 | 1.29 | 1.99 | 1.12 | 1.3 | 1.57 | -1.3 |
| porphyrin-containing compound catabolic process | 1 | 2.0 | 1.05 | 2.09 | 1.06 | 1.46 | 2.34 | 1.12 |
| heme oxidation | 1 | 2.0 | 1.05 | 2.09 | 1.06 | 1.46 | 2.34 | 1.12 |
| phosphorus metabolic process | 462 | 1.19 | 1.08 | 1.21 | 1.06 | 1.23 | 1.36 | 1.08 |
| phosphate-containing compound metabolic process | 462 | 1.19 | 1.08 | 1.21 | 1.06 | 1.23 | 1.36 | 1.08 |
| reactive oxygen species metabolic process | 13 | 1.12 | 1.07 | -1.12 | 1.06 | -1.02 | 1.06 | -1.05 |
| superoxide metabolic process | 7 | 1.3 | 1.1 | -1.14 | 1.03 | -1.08 | 1.27 | -1.04 |
| xenobiotic metabolic process | 10 | 1.1 | -1.02 | -1.79 | -1.06 | -1.28 | -1.46 | 1.06 |
| response to insecticide | 16 | 1.05 | 1.09 | -1.54 | -1.14 | -1.27 | -1.41 | 1.29 |
| nitrogen compound metabolic process | 1275 | 1.09 | 1.03 | 1.06 | 1.06 | 1.13 | 1.1 | 1.06 |
| nitric oxide biosynthetic process | 1 | 1.08 | 1.06 | 1.02 | 1.1 | 1.05 | -1.07 | -1.02 |
| transport | 1327 | -1.07 | 1.01 | 1.01 | -1.01 | 1.06 | 1.08 | 1.01 |
| ion transport | 298 | -1.06 | -1.04 | 1.01 | -1.1 | -1.05 | 1.19 | -1.05 |
| cation transport | 213 | -1.06 | -1.03 | 1.05 | -1.06 | -1.02 | 1.21 | -1.05 |
| potassium ion transport | 37 | 1.08 | -1.04 | 1.02 | -1.19 | -1.13 | 1.09 | -1.11 |
| sodium ion transport | 50 | -1.09 | -1.07 | -1.07 | -1.01 | -1.03 | 1.13 | -1.05 |
| calcium ion transport | 24 | 1.54 | -1.04 | 1.07 | -1.11 | 1.19 | 1.63 | 1.19 |
| phosphate ion transport | 10 | -1.16 | -1.04 | -1.52 | -1.15 | 1.01 | 2.17 | -1.68 |
| hydrogen transport | 51 | -1.18 | 1.0 | 1.25 | -1.1 | -1.04 | 1.38 | -1.15 |
| anion transport | 43 | -1.18 | -1.09 | -1.23 | -1.24 | -1.09 | 1.32 | -1.06 |
| chloride transport | 6 | 1.12 | 1.13 | -1.09 | 1.06 | 1.34 | 1.82 | 1.32 |
| metal ion transport | 132 | 1.02 | -1.06 | 1.01 | -1.06 | -1.03 | 1.18 | -1.03 |
| water transport | 3 | -1.65 | 1.03 | -1.57 | 1.34 | -1.36 | -2.48 | 1.11 |
| dicarboxylic acid transport | 8 | 1.09 | -1.04 | -1.41 | -1.66 | -1.35 | 1.44 | 1.09 |
| neurotransmitter transport | 121 | -1.01 | -1.02 | 1.09 | 1.04 | 1.22 | 1.22 | 1.08 |
| serotonin transport | 1 | -1.06 | -1.11 | -1.0 | -1.08 | -1.15 | 3.37 | -1.04 |
| mitochondrial transport | 49 | 1.06 | 1.1 | 1.12 | 1.12 | 1.18 | 1.27 | -1.05 |
| oxoglutarate:malate antiporter activity | 4 | -1.11 | -1.07 | 1.09 | -1.04 | -1.13 | 1.37 | -1.22 |
| alpha-ketoglutarate transport | 3 | -1.11 | -1.06 | 1.14 | -1.03 | -1.15 | 1.61 | -1.21 |
| malate transport | 5 | -1.06 | -1.22 | 1.26 | -1.12 | 1.12 | 2.23 | -1.26 |
| tricarboxylic acid transport | 6 | 1.38 | 1.1 | -1.07 | -1.11 | -1.31 | 1.27 | -1.04 |
| mitochondrial citrate transport | 1 | -1.29 | 1.86 | -1.38 | -1.29 | -2.25 | 1.02 | -1.13 |
| acyl carnitine transport | 2 | 1.34 | 1.32 | -1.91 | 1.22 | 1.14 | -1.7 | -1.23 |
| aspartate transport | 1 | 2.24 | 1.81 | -38.59 | -31.15 | -18.58 | -3.21 | 4.55 |
| L-glutamate transport | 4 | -2.92 | -1.1 | -4.98 | -1.98 | -1.32 | -2.35 | -3.75 |
| mitochondrial calcium ion transport | 1 | 1.01 | 1.3 | 1.51 | 1.08 | 2.35 | 4.41 | 1.77 |
| drug transmembrane transport | 1 | -10.37 | 1.6 | -11.08 | 1.04 | 2.2 | -3.48 | 1.28 |
| eye pigment precursor transport | 5 | -1.06 | 1.05 | 1.08 | -1.01 | 1.12 | -1.21 | 1.04 |
| oligopeptide transport | 3 | 1.69 | -1.17 | -1.51 | -1.15 | -1.07 | 1.17 | 1.2 |
| nucleotide transport | 3 | 1.1 | 1.07 | 1.13 | 1.06 | 1.12 | 1.41 | -1.2 |
| amino acid transport | 39 | -1.51 | -1.14 | -1.83 | -1.24 | 1.0 | -1.08 | -1.16 |
| glutamine transport | 1 | -3.47 | -4.26 | 1.06 | 2.44 | 2.17 | -3.37 | -7.09 |
| lipid transport | 28 | -2.26 | -1.6 | -1.93 | -1.01 | -1.22 | -1.56 | -1.89 |
| cellular ion homeostasis | 52 | 1.12 | 1.02 | 1.16 | -1.19 | -1.12 | 1.37 | 1.11 |
| cellular calcium ion homeostasis | 10 | 1.86 | -1.02 | 1.36 | -1.1 | -1.04 | 1.64 | 1.18 |
| cellular metal ion homeostasis | 20 | 1.2 | -1.1 | 1.18 | -1.09 | -1.21 | 1.18 | -1.03 |
| cellular copper ion homeostasis | 2 | -1.26 | 1.35 | 2.1 | 1.13 | 1.14 | 1.39 | 1.07 |
| cellular iron ion homeostasis | 8 | 1.49 | -1.19 | -1.16 | -1.17 | -1.18 | 1.48 | 1.09 |
| cellular zinc ion homeostasis | 1 | -1.15 | 1.5 | 3.36 | 1.09 | -1.02 | -1.02 | -1.03 |
| cell volume homeostasis | 2 | 1.92 | 1.08 | -1.06 | -1.08 | 2.11 | 2.25 | 1.09 |
| regulation of pH | 9 | -1.27 | 1.25 | 1.91 | -1.01 | 1.19 | 1.86 | 1.45 |
| intracellular protein transport | 172 | 1.02 | 1.18 | 1.29 | 1.13 | 1.21 | 1.11 | 1.17 |
| exocytosis | 60 | -1.02 | 1.08 | 1.27 | 1.11 | 1.21 | 1.16 | 1.11 |
| ER to Golgi vesicle-mediated transport | 17 | 1.16 | 1.15 | 1.31 | 1.14 | 1.22 | 1.36 | 1.31 |
| retrograde vesicle-mediated transport, Golgi to ER | 8 | -1.71 | 1.25 | -1.07 | 1.15 | -1.15 | -1.33 | 1.34 |
| intra-Golgi vesicle-mediated transport | 18 | -1.13 | 1.2 | 1.21 | 1.08 | 1.18 | 1.32 | 1.58 |
| post-Golgi vesicle-mediated transport | 8 | -1.19 | 1.11 | 1.49 | 1.14 | 1.49 | -1.0 | 1.24 |
| Golgi to plasma membrane transport | 1 | 1.1 | -1.05 | 1.04 | -1.06 | 1.01 | -1.16 | 1.25 |
| Golgi to endosome transport | 2 | 1.21 | 1.22 | 1.46 | 1.16 | 1.27 | -1.02 | 1.39 |
| Golgi to vacuole transport | 1 | 1.54 | 1.27 | 1.89 | 1.07 | 1.6 | 1.7 | 1.58 |
| endocytosis | 269 | 1.08 | 1.09 | 1.24 | 1.07 | 1.23 | 1.23 | 1.15 |
| receptor-mediated endocytosis | 11 | 1.09 | 1.13 | 1.29 | 1.01 | 1.19 | 1.55 | 1.26 |
| membrane budding | 17 | -1.05 | 1.2 | 1.38 | 1.12 | 1.21 | 1.27 | 1.27 |
| vesicle coating | 13 | 1.08 | 1.18 | 1.38 | 1.12 | 1.27 | 1.41 | 1.34 |
| vesicle docking involved in exocytosis | 27 | -1.1 | 1.16 | 1.4 | 1.15 | 1.26 | 1.14 | 1.1 |
| phagocytosis | 196 | 1.09 | 1.06 | 1.17 | 1.06 | 1.21 | 1.18 | 1.12 |
| phagocytosis, recognition | 1 | 1.05 | 1.01 | 1.06 | 1.12 | 1.02 | -1.02 | -1.17 |
| phagocytosis, engulfment | 183 | 1.09 | 1.08 | 1.18 | 1.07 | 1.21 | 1.18 | 1.13 |
| nucleocytoplasmic transport | 70 | 1.17 | 1.16 | 1.34 | 1.19 | 1.33 | 1.24 | 1.11 |
| autophagy | 25 | 1.13 | 1.2 | 1.42 | 1.16 | 1.34 | 1.29 | 1.18 |
| anti-apoptosis | 18 | 1.77 | 1.02 | 1.32 | -1.0 | 1.46 | 1.51 | 1.52 |
| induction of apoptosis | 25 | 1.08 | 1.15 | 1.03 | 1.12 | 1.26 | -1.08 | 1.24 |
| activation of cysteine-type endopeptidase activity involved in apoptotic process | 13 | 1.0 | 1.15 | 1.25 | 1.21 | 1.45 | 1.21 | 1.07 |
| cellular component disassembly involved in apoptotic process | 4 | -1.16 | 1.19 | 1.65 | 1.21 | 1.51 | -1.05 | -1.11 |
| cleavage of lamin | 1 | -11.86 | 1.52 | 2.29 | 1.45 | 1.79 | -4.95 | -7.15 |
| cleavage of cytoskeletal proteins involved in apoptosis | 1 | -11.86 | 1.52 | 2.29 | 1.45 | 1.79 | -4.95 | -7.15 |
| cellular component movement | 320 | 1.09 | 1.09 | 1.24 | 1.05 | 1.25 | 1.16 | 1.13 |
| substrate-dependent cell migration | 3 | -1.11 | 1.15 | 1.36 | -1.0 | 1.31 | 1.01 | 1.09 |
| substrate-dependent cell migration, cell extension | 3 | -1.11 | 1.15 | 1.36 | -1.0 | 1.31 | 1.01 | 1.09 |
| chemotaxis | 189 | 1.1 | 1.08 | 1.23 | 1.01 | 1.28 | 1.28 | 1.28 |
| regulation of muscle contraction | 5 | 2.39 | -1.02 | -2.05 | -2.54 | -2.17 | 1.16 | 1.24 |
| smooth muscle contraction | 3 | 1.0 | 1.28 | 1.13 | -1.04 | 1.24 | 1.32 | 1.1 |
| regulation of smooth muscle contraction | 1 | 1.0 | -1.08 | -1.22 | 1.04 | -1.09 | -1.09 | -1.09 |
| striated muscle contraction | 4 | -1.17 | -1.12 | -1.49 | -1.32 | -1.09 | 1.16 | 1.78 |
| regulation of striated muscle contraction | 2 | 1.48 | 1.35 | -6.16 | -5.66 | -4.42 | -1.74 | 2.14 |
| response to mechanical stimulus | 14 | 1.05 | -1.27 | 1.01 | -1.36 | -1.07 | 1.88 | 1.17 |
| sensory perception of mechanical stimulus | 23 | 1.04 | 1.07 | 1.22 | 1.04 | 1.17 | 1.31 | 1.04 |
| cellular membrane fusion | 16 | -1.15 | 1.08 | 1.27 | 1.24 | 1.25 | 1.01 | -1.03 |
| syncytium formation | 27 | 1.01 | 1.06 | 1.27 | -1.02 | 1.21 | 1.27 | 1.14 |
| response to stress | 636 | 1.22 | -1.02 | 1.06 | -1.01 | 1.12 | 1.19 | 1.12 |
| inflammatory response | 3 | -1.45 | 1.14 | 1.29 | 1.15 | 1.43 | -1.36 | 1.17 |
| immune response | 137 | 1.38 | -1.24 | -1.13 | -1.19 | -1.03 | 1.17 | 1.17 |
| humoral immune response | 72 | 1.45 | -1.32 | -1.21 | -1.31 | -1.05 | 1.33 | 1.25 |
| male-specific antibacterial humoral response | 1 | -1.1 | -1.03 | -1.03 | -1.0 | -1.23 | -1.2 | -1.1 |
| pattern orientation | 1 | 1.15 | 1.52 | 1.62 | 1.11 | 1.74 | 1.53 | 2.02 |
| positive regulation of antibacterial peptide biosynthetic process | 17 | 1.54 | -1.16 | -1.04 | -1.14 | 1.02 | 1.29 | 1.37 |
| positive regulation of biosynthetic process of antibacterial peptides active against Gram-negative bacteria | 5 | 2.54 | -1.22 | -1.03 | -1.31 | -1.06 | 2.04 | 1.48 |
| positive regulation of biosynthetic process of antibacterial peptides active against Gram-positive bacteria | 4 | 1.05 | -1.9 | -2.24 | -1.85 | -1.9 | -1.18 | 1.68 |
| positive regulation of antifungal peptide biosynthetic process | 6 | 2.45 | -1.42 | -1.16 | -1.01 | -1.05 | 1.77 | 1.2 |
| cellular defense response | 1 | 1.57 | 1.3 | 1.87 | 1.1 | 1.6 | 2.04 | 2.1 |
| response to osmotic stress | 8 | 1.24 | -1.08 | 1.33 | -1.08 | 1.22 | 1.12 | 1.39 |
| hyperosmotic response | 1 | 1.22 | 1.33 | 2.18 | 1.07 | 1.72 | 2.05 | 1.61 |
| response to DNA damage stimulus | 227 | 1.22 | 1.12 | 1.25 | 1.15 | 1.3 | 1.15 | 1.09 |
| response to oxidative stress | 70 | 1.24 | 1.01 | -1.04 | -1.06 | 1.02 | 1.38 | 1.1 |
| response to lipid hydroperoxide | 1 | 2.0 | -1.09 | -1.07 | 1.14 | 1.47 | 1.02 | -1.28 |
| ER overload response | 2 | -1.22 | 1.43 | 1.56 | 1.09 | 1.42 | -1.17 | 1.79 |
| ER-nucleus signaling pathway | 9 | 1.04 | 1.26 | 1.24 | 1.08 | 1.25 | -1.07 | 1.26 |
| positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response | 1 | -1.05 | 1.27 | -1.17 | -1.24 | -1.49 | -1.01 | 1.19 |
| response to sterol depletion | 1 | 1.18 | 1.17 | 1.43 | 1.18 | 1.36 | -1.03 | 1.42 |
| positive regulation of transcription via sterol regulatory element binding involved in ER-nuclear sterol response pathway | 1 | 1.18 | 1.17 | 1.43 | 1.18 | 1.36 | -1.03 | 1.42 |
| organelle organization | 973 | 1.15 | 1.09 | 1.25 | 1.09 | 1.27 | 1.18 | 1.1 |
| nucleus organization | 36 | 1.03 | 1.15 | 1.23 | 1.12 | 1.36 | 1.0 | 1.13 |
| nuclear envelope organization | 6 | 1.25 | 1.18 | 1.06 | 1.25 | 1.16 | -1.19 | -1.0 |
| nuclear pore organization | 1 | 1.8 | 1.61 | 2.39 | 1.2 | 2.33 | 2.16 | 2.46 |
| nucleolus organization | 3 | 1.16 | 1.19 | 1.66 | 1.24 | 2.3 | 1.1 | 1.42 |
| mitochondrion organization | 74 | 1.04 | 1.08 | 1.25 | 1.08 | 1.18 | 1.25 | -1.14 |
| mitochondrial membrane organization | 10 | 1.09 | 1.08 | 1.26 | 1.13 | 1.21 | 1.18 | -1.04 |
| inner mitochondrial membrane organization | 7 | 1.09 | 1.08 | 1.28 | 1.16 | 1.18 | 1.3 | -1.13 |
| plasma membrane organization | 12 | 1.23 | 1.13 | 1.5 | 1.02 | 1.84 | 1.37 | 1.27 |
| actin filament organization | 87 | 1.01 | 1.07 | 1.38 | 1.03 | 1.32 | 1.12 | 1.17 |
| cytoskeletal anchoring at plasma membrane | 17 | 1.37 | 1.1 | 1.43 | 1.03 | 1.41 | 1.51 | 1.32 |
| microtubule-based process | 438 | 1.12 | 1.09 | 1.22 | 1.08 | 1.22 | 1.17 | 1.08 |
| microtubule-based movement | 107 | -1.02 | 1.09 | 1.13 | 1.07 | 1.1 | 1.04 | -1.01 |
| microtubule depolymerization | 3 | -1.13 | 1.2 | 1.15 | 1.08 | 1.3 | 1.07 | 1.1 |
| microtubule nucleation | 7 | 1.67 | 1.03 | 1.42 | 1.09 | 1.45 | 1.62 | 1.17 |
| tubulin complex assembly | 4 | 1.19 | 1.1 | 1.33 | 1.15 | 1.38 | -1.02 | 1.05 |
| negative regulation of microtubule depolymerization | 15 | 1.06 | 1.15 | 1.48 | 1.04 | 1.41 | 1.59 | 1.26 |
| cytoplasm organization | 51 | 1.09 | -1.03 | 1.09 | -1.08 | 1.21 | 1.39 | 1.18 |
| endoplasmic reticulum organization | 7 | 1.12 | 1.24 | 1.29 | 1.1 | 1.28 | 1.04 | 1.52 |
| Golgi organization | 47 | 1.19 | 1.1 | 1.16 | 1.09 | 1.17 | 1.1 | 1.13 |
| peroxisome organization | 15 | 1.58 | 1.07 | -1.39 | 1.25 | 1.36 | -1.44 | -1.19 |
| endosome organization | 5 | 1.15 | 1.16 | 1.31 | 1.2 | 1.26 | 1.15 | 1.21 |
| vacuolar transport | 17 | 1.14 | 1.17 | 1.78 | 1.13 | 1.5 | 1.47 | 1.29 |
| vacuolar acidification | 4 | -2.16 | 1.36 | 2.14 | -1.07 | 1.21 | 2.24 | 1.25 |
| lysosome organization | 10 | -1.08 | 1.12 | 1.05 | 1.24 | 1.07 | -1.05 | 1.03 |
| lysosomal transport | 13 | -1.01 | 1.15 | 1.71 | 1.13 | 1.47 | 1.31 | 1.22 |
| cell-cell junction assembly | 36 | 1.91 | -1.1 | 1.31 | -1.15 | 1.22 | 2.16 | 1.42 |
| cell-substrate junction assembly | 1 | -1.1 | -1.15 | -1.07 | -1.18 | 1.52 | 1.28 | 1.24 |
| cell cycle | 573 | 1.19 | 1.11 | 1.23 | 1.11 | 1.27 | 1.19 | 1.1 |
| cell cycle arrest | 8 | 1.45 | 1.2 | 1.34 | 1.1 | 1.36 | 1.5 | 1.38 |
| spindle organization | 213 | 1.2 | 1.08 | 1.21 | 1.07 | 1.26 | 1.2 | 1.09 |
| mitotic spindle organization | 189 | 1.21 | 1.09 | 1.21 | 1.1 | 1.26 | 1.18 | 1.06 |
| spindle assembly involved in male meiosis | 3 | -1.71 | 1.36 | 1.08 | 1.16 | 1.08 | -1.71 | 1.01 |
| spindle assembly involved in male meiosis I | 1 | 1.11 | 1.51 | 1.01 | 1.21 | 1.06 | -1.41 | 1.26 |
| spindle assembly involved in female meiosis | 11 | 1.05 | 1.02 | 1.05 | 1.05 | 1.26 | -1.08 | -1.04 |
| spindle assembly involved in female meiosis I | 3 | 1.06 | -1.05 | -1.08 | -1.02 | 1.25 | -1.28 | -1.07 |
| spindle assembly involved in female meiosis II | 4 | 1.32 | 1.05 | 1.25 | 1.09 | 1.47 | 1.13 | 1.04 |
| male meiosis chromosome segregation | 9 | 1.31 | 1.09 | 1.3 | 1.08 | 1.38 | 1.2 | 1.09 |
| sister chromatid cohesion | 20 | 1.2 | 1.1 | 1.27 | 1.08 | 1.29 | 1.25 | 1.11 |
| mitotic sister chromatid cohesion | 5 | 1.6 | 1.06 | 1.64 | 1.1 | 1.59 | 1.67 | 1.28 |
| male meiosis sister chromatid cohesion | 4 | -1.07 | 1.07 | 1.03 | 1.03 | 1.12 | -1.1 | -1.1 |
| female meiosis sister chromatid cohesion | 1 | 1.04 | 1.22 | 1.16 | 1.15 | 1.04 | -1.0 | -1.09 |
| mitosis | 144 | 1.21 | 1.1 | 1.27 | 1.12 | 1.33 | 1.24 | 1.1 |
| negative regulation of transcription during mitosis | 4 | 1.21 | 1.13 | 1.22 | 1.16 | 1.17 | 1.14 | 1.08 |
| negative regulation of transcription from RNA polymerase II promoter during mitosis | 4 | 1.21 | 1.13 | 1.22 | 1.16 | 1.17 | 1.14 | 1.08 |
| mitotic chromosome condensation | 20 | 1.29 | 1.08 | 1.28 | 1.1 | 1.34 | 1.37 | 1.15 |
| mitotic chromosome movement towards spindle pole | 1 | 1.43 | 1.0 | -1.06 | 1.15 | 1.3 | -1.5 | -1.26 |
| mitotic metaphase plate congression | 13 | 1.3 | 1.03 | 1.22 | 1.18 | 1.42 | 1.19 | -1.01 |
| mitotic nuclear envelope reassembly | 2 | 1.28 | 1.06 | -1.0 | 1.32 | 1.15 | -1.17 | -1.1 |
| regulation of mitosis | 45 | 1.26 | 1.07 | 1.15 | 1.07 | 1.27 | 1.23 | 1.12 |
| regulation of S phase of mitotic cell cycle | 16 | 1.38 | 1.18 | 1.34 | 1.13 | 1.4 | 1.63 | 1.43 |
| mitotic cell cycle checkpoint | 76 | 1.22 | 1.15 | 1.34 | 1.18 | 1.38 | 1.28 | 1.15 |
| mitotic cell cycle spindle assembly checkpoint | 10 | 1.07 | 1.09 | 1.16 | 1.15 | 1.32 | 1.1 | 1.03 |
| optokinetic behavior | 2 | 1.16 | 1.09 | 1.72 | 1.19 | 1.37 | 1.2 | 2.11 |
| mitotic cell cycle G2/M transition DNA damage checkpoint | 62 | 1.22 | 1.15 | 1.36 | 1.19 | 1.38 | 1.27 | 1.14 |
| regulation of exit from mitosis | 9 | 1.33 | 1.04 | 1.03 | 1.01 | 1.18 | 1.35 | -1.03 |
| nuclear migration | 21 | 1.07 | 1.19 | 1.32 | 1.14 | 1.34 | 1.16 | 1.17 |
| centrosome cycle | 87 | 1.19 | 1.09 | 1.2 | 1.08 | 1.26 | 1.16 | 1.1 |
| centriole replication | 18 | 1.17 | 1.08 | 1.22 | 1.05 | 1.29 | 1.15 | 1.1 |
| mitotic centrosome separation | 5 | -1.07 | 1.01 | -1.06 | 1.11 | 1.17 | -1.05 | -1.17 |
| cytokinesis, completion of separation | 1 | -1.24 | -1.05 | 1.11 | 1.18 | 1.4 | -1.36 | -1.18 |
| cytokinesis after meiosis I | 8 | 1.52 | -1.23 | 1.02 | -1.47 | 1.08 | 1.67 | 1.38 |
| cytokinesis after meiosis II | 7 | 1.71 | -1.31 | -1.08 | -1.6 | 1.08 | 2.04 | 1.46 |
| male meiosis cytokinesis | 14 | 1.82 | -1.04 | 1.11 | -1.11 | 1.39 | 1.32 | 1.33 |
| endomitotic cell cycle | 9 | 1.39 | 1.23 | 1.35 | 1.28 | 1.47 | 1.22 | 1.19 |
| establishment of cell polarity | 27 | 1.11 | 1.12 | 1.49 | 1.04 | 1.4 | 1.27 | 1.22 |
| meiosis | 124 | 1.1 | 1.11 | 1.17 | 1.11 | 1.22 | 1.09 | 1.08 |
| meiosis I | 52 | 1.05 | 1.13 | 1.13 | 1.1 | 1.19 | 1.04 | 1.05 |
| meiotic prophase I | 1 | 1.5 | 1.02 | 1.68 | -1.05 | 1.78 | 1.86 | 1.49 |
| synapsis | 2 | 1.33 | 1.05 | 1.23 | 1.11 | 1.31 | -1.03 | 1.07 |
| synaptonemal complex assembly | 1 | 1.0 | 1.01 | -1.04 | 1.07 | -1.07 | -1.07 | -1.01 |
| reciprocal meiotic recombination | 22 | 1.06 | 1.12 | 1.07 | 1.13 | 1.17 | 1.01 | 1.01 |
| meiotic metaphase I | 1 | 1.38 | 1.21 | 1.33 | 1.18 | 1.64 | 1.12 | 1.06 |
| meiotic anaphase I | 4 | 1.0 | 1.05 | 1.11 | 1.01 | 1.1 | 1.09 | 1.06 |
| meiosis II | 8 | 1.18 | 1.02 | 1.12 | 1.03 | 1.24 | 1.13 | 1.02 |
| meiotic anaphase II | 2 | 1.2 | 1.05 | 1.15 | 1.01 | 1.2 | 1.43 | 1.16 |
| male meiosis | 41 | -1.02 | 1.14 | 1.17 | 1.1 | 1.19 | -1.06 | 1.05 |
| male meiosis I | 8 | -1.07 | 1.2 | 1.15 | 1.06 | 1.14 | -1.22 | 1.12 |
| female meiosis | 61 | 1.13 | 1.07 | 1.17 | 1.1 | 1.24 | 1.12 | 1.07 |
| female meiosis I | 6 | 1.01 | -1.05 | -1.08 | -1.0 | 1.15 | -1.17 | -1.1 |
| meiotic recombination nodule assembly | 1 | -1.21 | 1.44 | 1.1 | 1.2 | 1.03 | -1.18 | 1.17 |
| female meiosis II | 6 | 1.18 | 1.01 | 1.11 | 1.04 | 1.25 | 1.04 | -1.03 |
| growth | 135 | 1.26 | 1.12 | 1.23 | -1.01 | 1.26 | 1.34 | 1.29 |
| cell communication | 1251 | 1.14 | 1.03 | 1.15 | 1.03 | 1.17 | 1.18 | 1.11 |
| homophilic cell adhesion | 27 | 1.7 | -1.16 | -1.1 | -1.17 | 1.12 | 1.92 | 1.39 |
| neuron cell-cell adhesion | 1 | 1.45 | 1.31 | 1.24 | 1.07 | 1.17 | 1.33 | 1.2 |
| cell-matrix adhesion | 16 | -1.41 | 1.01 | 1.24 | -1.06 | 1.3 | -1.06 | 1.21 |
| negative regulation of cell adhesion | 9 | 1.95 | 1.17 | 1.32 | 1.03 | 1.28 | 1.88 | 1.51 |
| establishment or maintenance of cell polarity | 152 | 1.11 | 1.11 | 1.36 | 1.05 | 1.29 | 1.3 | 1.31 |
| establishment of tissue polarity | 70 | 1.2 | 1.06 | 1.23 | 1.01 | 1.28 | 1.24 | 1.46 |
| cell surface receptor signaling pathway | 482 | 1.17 | -1.01 | 1.15 | -1.01 | 1.13 | 1.17 | 1.12 |
| enzyme linked receptor protein signaling pathway | 151 | 1.22 | 1.07 | 1.3 | 1.05 | 1.25 | 1.29 | 1.24 |
| signal complex assembly | 1 | 1.01 | 1.19 | 1.87 | 1.24 | 1.92 | 1.43 | 1.32 |
| epidermal growth factor receptor signaling pathway | 34 | 1.44 | 1.08 | 1.27 | 1.08 | 1.29 | 1.29 | 1.22 |
| regulation of epidermal growth factor-activated receptor activity | 4 | 1.16 | 1.09 | 1.49 | 1.02 | 1.37 | 1.48 | 1.2 |
| transforming growth factor beta receptor signaling pathway | 20 | 1.22 | 1.04 | 1.25 | -1.09 | 1.26 | 1.29 | 1.17 |
| transforming growth factor beta binding | 1 | -2.53 | 1.23 | -1.01 | 1.28 | 2.46 | 1.22 | 1.76 |
| transforming growth factor beta receptor complex assembly | 1 | -2.53 | 1.23 | -1.01 | 1.28 | 2.46 | 1.22 | 1.76 |
| SMAD protein import into nucleus | 12 | -1.03 | 1.3 | 1.39 | 1.26 | 1.48 | 1.18 | 1.22 |
| transmembrane receptor protein tyrosine phosphatase signaling pathway | 1 | 1.65 | 1.02 | 2.09 | -1.05 | 1.83 | 4.08 | 1.95 |
| G-protein coupled receptor signaling pathway | 176 | 1.02 | -1.09 | 1.05 | -1.05 | 1.0 | -1.09 | -1.02 |
| G-protein signaling, coupled to cyclic nucleotide second messenger | 6 | 1.08 | 1.08 | 1.0 | 1.05 | -1.0 | -1.03 | -1.04 |
| G-protein signaling, coupled to cAMP nucleotide second messenger | 6 | 1.08 | 1.08 | 1.0 | 1.05 | -1.0 | -1.03 | -1.04 |
| activation of adenylate cyclase activity by G-protein signaling pathway | 3 | -1.05 | 1.08 | -1.04 | 1.0 | -1.08 | -1.13 | -1.16 |
| activation of adenylate cyclase activity | 5 | 1.21 | 1.07 | 1.13 | 1.0 | 1.06 | 1.08 | -1.01 |
| activation of adenylate cyclase activity by dopamine receptor signaling pathway | 2 | -1.21 | 1.11 | -1.21 | 1.0 | -1.21 | -1.22 | -1.28 |
| activation of adenylate cyclase activity by serotonin receptor signaling pathway | 1 | 1.27 | 1.02 | 1.28 | 1.01 | 1.16 | 1.03 | 1.05 |
| inhibition of adenylate cyclase activity by G-protein signaling pathway | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| negative regulation of adenylate cyclase activity | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| inhibition of adenylate cyclase activity by serotonin receptor signaling pathway | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| activation of phospholipase C activity by G-protein coupled receptor protein signaling pathway coupled to IP3 second messenger | 4 | 1.12 | 1.13 | 1.06 | 1.07 | 1.06 | 1.07 | 1.09 |
| activation of phospholipase C activity | 6 | 1.13 | 1.12 | 1.17 | 1.04 | 1.09 | 1.14 | 1.05 |
| elevation of cytosolic calcium ion concentration | 3 | 1.46 | 1.16 | 1.52 | -1.0 | 1.18 | 1.77 | 1.54 |
| activation of protein kinase C activity by G-protein coupled receptor protein signaling pathway | 8 | 1.65 | 1.05 | -1.03 | -1.02 | -1.04 | 1.17 | 1.24 |
| activation of phospholipase C activity by serotonin receptor signaling pathway | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| serotonin receptor signaling pathway | 3 | 1.27 | -1.02 | 1.07 | 1.0 | 1.01 | 1.11 | 1.03 |
| octopamine or tyramine signaling pathway | 4 | -1.04 | 1.16 | -1.05 | 1.12 | 1.02 | -1.17 | 1.09 |
| dopamine receptor signaling pathway | 3 | -1.16 | 1.05 | -1.23 | 1.04 | -1.15 | -1.24 | -1.21 |
| G-protein coupled acetylcholine receptor signaling pathway | 2 | 1.0 | 1.08 | 1.1 | 1.16 | 1.17 | 1.06 | 1.05 |
| gamma-aminobutyric acid signaling pathway | 1 | -1.12 | -1.07 | -1.06 | -1.08 | -1.01 | 1.07 | -1.02 |
| glutamate receptor signaling pathway | 5 | -1.03 | 1.1 | 1.0 | 1.09 | -1.01 | 1.02 | 1.02 |
| G-protein coupled glutamate receptor signaling pathway | 3 | 1.11 | 1.14 | 1.05 | 1.15 | 1.03 | 1.07 | 1.0 |
| tachykinin receptor signaling pathway | 5 | -1.04 | -1.19 | 1.08 | -1.24 | -1.02 | 1.95 | 1.43 |
| neuropeptide signaling pathway | 38 | -1.86 | -1.39 | -1.2 | -1.15 | -1.16 | -1.83 | -1.26 |
| Notch signaling pathway | 41 | -1.09 | 1.14 | 1.18 | 1.13 | 1.3 | 1.07 | 1.15 |
| Notch receptor processing | 4 | -1.0 | 1.17 | 1.21 | 1.25 | 1.56 | 1.22 | 1.41 |
| smoothened signaling pathway | 30 | 1.48 | 1.03 | 1.07 | -1.08 | 1.17 | 1.49 | 1.16 |
| patched ligand maturation | 1 | 1.07 | 1.32 | 1.75 | 1.48 | 1.85 | -1.07 | -1.03 |
| signal transduction downstream of smoothened | 5 | 1.03 | 1.18 | 1.56 | 1.12 | 1.42 | 1.4 | 1.07 |
| positive regulation of hh target transcription factor activity | 5 | 1.03 | 1.18 | 1.56 | 1.12 | 1.42 | 1.4 | 1.07 |
| integrin-mediated signaling pathway | 4 | -1.54 | 1.07 | 1.65 | 1.1 | 1.77 | -1.22 | 1.11 |
| calcium-mediated signaling | 8 | -1.04 | 1.02 | -1.11 | 1.01 | -1.17 | 1.07 | -1.03 |
| intracellular protein kinase cascade | 51 | -1.02 | 1.16 | 1.44 | 1.09 | 1.44 | 1.16 | 1.28 |
| I-kappaB kinase/NF-kappaB cascade | 2 | 1.26 | 1.28 | 1.33 | 1.08 | 1.48 | 1.61 | 1.39 |
| JNK cascade | 25 | 1.0 | 1.14 | 1.47 | 1.1 | 1.5 | 1.24 | 1.42 |
| activation of JNKK activity | 2 | 1.23 | 1.27 | 1.56 | 1.2 | 1.54 | 1.23 | 1.3 |
| activation of JUN kinase activity | 2 | -1.08 | 1.31 | 1.72 | 1.2 | 1.5 | 1.32 | 1.54 |
| JUN phosphorylation | 1 | -1.04 | -1.12 | 1.46 | 1.28 | 1.4 | -1.15 | 1.1 |
| JAK-STAT cascade | 11 | -1.2 | 1.17 | 1.35 | 1.1 | 1.37 | -1.26 | 1.18 |
| tyrosine phosphorylation of STAT protein | 1 | 1.57 | 1.3 | 1.87 | 1.1 | 1.6 | 2.04 | 2.1 |
| STAT protein import into nucleus | 1 | 1.57 | 1.3 | 1.87 | 1.1 | 1.6 | 2.04 | 2.1 |
| small GTPase mediated signal transduction | 105 | 1.17 | 1.12 | 1.35 | 1.08 | 1.3 | 1.17 | 1.18 |
| Ras protein signal transduction | 25 | 1.08 | 1.28 | 1.57 | 1.08 | 1.47 | 1.22 | 1.49 |
| Rho protein signal transduction | 4 | -1.15 | 1.27 | 1.54 | 1.2 | 1.45 | 1.01 | 1.39 |
| cell-cell signaling | 419 | 1.13 | 1.06 | 1.16 | 1.07 | 1.2 | 1.18 | 1.08 |
| synaptic transmission | 216 | 1.09 | 1.08 | 1.21 | 1.04 | 1.19 | 1.18 | 1.13 |
| neurotransmitter secretion | 99 | -1.06 | 1.05 | 1.24 | 1.12 | 1.25 | 1.11 | 1.05 |
| neuron-neuron synaptic transmission | 5 | 1.1 | -1.05 | -1.09 | -1.15 | 1.01 | 2.0 | 1.04 |
| synaptic transmission, cholinergic | 1 | 2.27 | 1.23 | -5.17 | -1.59 | -2.2 | -9.9 | 9.42 |
| ensheathment of neurons | 6 | 1.77 | -1.07 | 1.38 | -1.13 | 1.24 | 3.0 | 1.46 |
| neuromuscular synaptic transmission | 27 | 1.22 | 1.11 | 1.27 | 1.0 | 1.12 | 1.31 | 1.11 |
| multicellular organismal development | 2108 | 1.15 | 1.05 | 1.17 | 1.04 | 1.19 | 1.21 | 1.13 |
| gamete generation | 583 | 1.14 | 1.1 | 1.22 | 1.05 | 1.25 | 1.21 | 1.19 |
| pole cell development | 21 | 1.16 | 1.17 | 1.47 | 1.09 | 1.43 | 1.28 | 1.24 |
| pole cell fate determination | 3 | 2.8 | 1.18 | 2.04 | 1.19 | 1.51 | 2.12 | 1.41 |
| pole cell formation | 17 | 1.13 | 1.16 | 1.33 | 1.07 | 1.3 | 1.17 | 1.22 |
| pole cell migration | 19 | 1.23 | -1.05 | 1.15 | 1.01 | 1.33 | 1.36 | 1.04 |
| germ cell development | 228 | 1.1 | 1.14 | 1.29 | 1.08 | 1.3 | 1.25 | 1.19 |
| cystoblast division | 9 | 1.07 | 1.12 | 1.19 | 1.15 | 1.29 | 1.17 | 1.17 |
| spermatogenesis | 150 | 1.07 | 1.14 | 1.22 | 1.07 | 1.24 | 1.12 | 1.11 |
| spermatogonial cell division | 5 | -1.04 | 1.03 | 1.09 | 1.21 | 1.28 | -1.18 | -1.02 |
| primary spermatocyte growth | 5 | 2.0 | 1.25 | 1.67 | 1.27 | 1.92 | 1.51 | 1.27 |
| spermatid development | 79 | 1.04 | 1.12 | 1.3 | 1.04 | 1.29 | 1.22 | 1.17 |
| Nebenkern assembly | 2 | 1.05 | 1.03 | 1.07 | -1.02 | 1.14 | 1.17 | 1.07 |
| sperm axoneme assembly | 7 | -1.01 | 1.09 | 1.1 | 1.07 | 1.15 | -1.0 | 1.02 |
| spermatid nucleus differentiation | 9 | -1.34 | 1.07 | 1.03 | 1.03 | 1.26 | -1.23 | 1.02 |
| spermatid nucleus elongation | 2 | 1.34 | 1.05 | 1.39 | 1.01 | 1.37 | 1.32 | 1.14 |
| sperm individualization | 44 | -1.12 | 1.16 | 1.4 | 1.06 | 1.35 | 1.23 | 1.23 |
| female gamete generation | 427 | 1.13 | 1.08 | 1.22 | 1.04 | 1.24 | 1.22 | 1.2 |
| germarium-derived egg chamber formation | 61 | 1.02 | 1.14 | 1.23 | 1.1 | 1.31 | 1.21 | 1.23 |
| germarium-derived oocyte fate determination | 21 | 1.11 | 1.14 | 1.23 | 1.09 | 1.21 | 1.3 | 1.23 |
| growth of a germarium-derived egg chamber | 5 | 1.5 | 1.13 | 1.67 | 1.04 | 1.5 | 1.59 | 1.41 |
| vitellogenesis | 7 | 1.4 | -3.65 | -4.11 | -4.07 | -1.34 | 2.21 | 1.12 |
| ovarian follicle cell migration | 79 | 1.03 | 1.16 | 1.44 | 1.13 | 1.47 | 1.15 | 1.19 |
| border follicle cell migration | 72 | 1.05 | 1.17 | 1.45 | 1.13 | 1.49 | 1.2 | 1.21 |
| ovarian follicle cell-cell adhesion | 3 | -1.5 | 1.41 | 1.37 | 1.22 | 1.72 | -1.12 | -1.09 |
| ovarian nurse cell to oocyte transport | 32 | -1.11 | 1.14 | 1.27 | 1.11 | 1.3 | -1.14 | 1.1 |
| female germline ring canal formation | 9 | -1.09 | 1.12 | 1.17 | 1.11 | 1.32 | 1.12 | 1.24 |
| nurse cell nucleus anchoring | 1 | -1.33 | 1.57 | 2.09 | 1.17 | 1.77 | 1.11 | 1.58 |
| cytoplasmic transport, nurse cell to oocyte | 17 | -1.19 | 1.13 | 1.2 | 1.1 | 1.19 | -1.42 | -1.04 |
| chorion-containing eggshell formation | 78 | 1.18 | 1.1 | 1.22 | 1.06 | 1.23 | 1.11 | 1.17 |
| vitelline membrane formation involved in chorion-containing eggshell formation | 12 | 1.14 | 1.01 | -1.05 | -1.02 | -1.08 | 1.01 | -1.0 |
| eggshell chorion assembly | 60 | 1.26 | 1.11 | 1.27 | 1.08 | 1.27 | 1.14 | 1.2 |
| eggshell chorion gene amplification | 17 | 1.11 | 1.21 | 1.43 | 1.19 | 1.52 | 1.14 | 1.14 |
| oocyte construction | 96 | 1.16 | 1.13 | 1.34 | 1.07 | 1.27 | 1.39 | 1.25 |
| oocyte axis specification | 93 | 1.19 | 1.13 | 1.33 | 1.07 | 1.25 | 1.4 | 1.25 |
| oocyte dorsal/ventral axis specification | 34 | 1.43 | 1.04 | 1.13 | 1.01 | 1.1 | 1.5 | 1.28 |
| maternal specification of dorsal/ventral axis, oocyte, germ-line encoded | 7 | 1.96 | -1.07 | -1.01 | -1.08 | 1.03 | 2.21 | 1.39 |
| oocyte nucleus migration involved in oocyte dorsal/ventral axis specification | 5 | 1.19 | 1.19 | 1.21 | 1.12 | 1.21 | 1.46 | 1.07 |
| maternal specification of dorsal/ventral axis, oocyte, soma encoded | 6 | 1.43 | -1.12 | 1.09 | -1.23 | -1.1 | -1.09 | 1.5 |
| oocyte anterior/posterior axis specification | 62 | 1.11 | 1.18 | 1.4 | 1.13 | 1.34 | 1.37 | 1.21 |
| pole plasm assembly | 44 | 1.05 | 1.19 | 1.38 | 1.14 | 1.31 | 1.29 | 1.19 |
| pole plasm RNA localization | 37 | 1.02 | 1.19 | 1.31 | 1.15 | 1.26 | 1.21 | 1.17 |
| regulation of pole plasm oskar mRNA localization | 25 | -1.02 | 1.16 | 1.34 | 1.11 | 1.28 | 1.31 | 1.17 |
| pole plasm protein localization | 10 | 1.29 | 1.15 | 1.44 | 1.11 | 1.37 | 1.64 | 1.23 |
| negative regulation of oskar mRNA translation | 8 | 1.19 | -1.01 | 1.28 | -1.09 | 1.13 | 1.88 | 1.01 |
| insemination | 17 | 1.27 | -1.21 | -1.24 | -1.29 | -1.17 | 1.36 | -1.21 |
| sperm displacement | 3 | 1.07 | 1.02 | 1.02 | 1.02 | 1.05 | -1.05 | -1.0 |
| single fertilization | 21 | 1.03 | 1.13 | 1.12 | 1.07 | 1.1 | 1.04 | 1.03 |
| egg activation | 9 | 1.11 | 1.13 | -1.02 | 1.02 | -1.1 | 1.13 | 1.03 |
| pronuclear fusion | 6 | 1.05 | 1.11 | 1.23 | 1.09 | 1.44 | 1.02 | 1.13 |
| anatomical structure morphogenesis | 1160 | 1.14 | 1.06 | 1.19 | 1.02 | 1.21 | 1.23 | 1.19 |
| embryo development | 457 | 1.16 | 1.04 | 1.23 | 1.01 | 1.25 | 1.28 | 1.21 |
| regulation of mitotic cell cycle | 169 | 1.21 | 1.12 | 1.24 | 1.12 | 1.31 | 1.26 | 1.14 |
| regulation of preblastoderm mitotic cell cycle | 1 | 2.87 | 1.19 | 1.15 | 1.16 | 1.39 | 3.16 | 1.43 |
| regulation of syncytial blastoderm mitotic cell cycle | 4 | -1.06 | 1.09 | 1.06 | 1.04 | -1.06 | -1.05 | -1.09 |
| cellularization | 88 | 1.03 | 1.13 | 1.34 | 1.07 | 1.31 | 1.26 | 1.23 |
| blastoderm segmentation | 173 | 1.2 | 1.1 | 1.26 | 1.04 | 1.23 | 1.26 | 1.24 |
| tripartite regional subdivision | 114 | 1.21 | 1.09 | 1.29 | 1.04 | 1.25 | 1.28 | 1.24 |
| zygotic specification of dorsal/ventral axis | 5 | 1.69 | 1.14 | 1.51 | 1.04 | 1.21 | 1.21 | 1.19 |
| embryonic pattern specification | 187 | 1.24 | 1.08 | 1.23 | 1.02 | 1.22 | 1.28 | 1.27 |
| zygotic determination of anterior/posterior axis, embryo | 31 | 1.57 | -1.1 | 1.04 | -1.14 | 1.04 | 1.27 | 1.27 |
| anterior region determination | 3 | 1.62 | -1.05 | 1.0 | -1.14 | -1.1 | 1.06 | -1.02 |
| posterior abdomen determination | 1 | 2.58 | 1.01 | 1.0 | 1.14 | -1.0 | -1.14 | 1.0 |
| terminal region determination | 24 | 1.44 | -1.08 | 1.07 | -1.1 | 1.09 | 1.37 | 1.32 |
| periodic partitioning | 36 | 1.13 | 1.1 | 1.13 | -1.03 | 1.27 | 1.27 | 1.33 |
| periodic partitioning by pair rule gene | 8 | 1.03 | 1.2 | -1.11 | -1.37 | 1.26 | 1.17 | 1.1 |
| segment polarity determination | 23 | 1.21 | 1.06 | 1.16 | 1.04 | 1.26 | 1.35 | 1.41 |
| determination of left/right symmetry | 4 | -3.48 | 1.34 | 1.25 | 1.01 | 1.41 | -1.67 | 1.12 |
| gastrulation | 69 | 1.19 | 1.03 | 1.19 | 1.02 | 1.24 | 1.29 | 1.11 |
| ventral furrow formation | 13 | 1.35 | 1.11 | 1.28 | 1.08 | 1.5 | 1.38 | 1.28 |
| posterior midgut invagination | 4 | 1.75 | 1.13 | 1.9 | 1.13 | 1.88 | 1.93 | 1.72 |
| anterior midgut invagination | 3 | 1.4 | 1.11 | 1.46 | 1.17 | 1.72 | 1.88 | 1.31 |
| cephalic furrow formation | 2 | 1.01 | 1.11 | 1.05 | -1.06 | 1.03 | 1.09 | 1.05 |
| germ-band extension | 8 | -1.08 | -1.25 | 1.17 | 1.08 | 1.55 | 1.32 | -1.22 |
| amnioserosa formation | 5 | 1.56 | -1.06 | 1.14 | -1.19 | -1.08 | -1.04 | 1.05 |
| segment specification | 32 | 1.1 | 1.02 | 1.19 | 1.11 | 1.15 | 1.21 | 1.01 |
| specification of segmental identity, head | 14 | 1.19 | 1.01 | 1.1 | 1.09 | -1.11 | -1.03 | -1.01 |
| specification of segmental identity, labial segment | 3 | 1.02 | 1.03 | 1.12 | 1.05 | -1.06 | 1.02 | 1.01 |
| specification of segmental identity, maxillary segment | 2 | 1.16 | 1.08 | 1.11 | 1.03 | 1.02 | -1.01 | -1.01 |
| specification of segmental identity, antennal segment | 3 | 1.59 | -1.13 | 1.32 | 1.09 | -1.18 | -1.05 | -1.16 |
| specification of segmental identity, thorax | 2 | 1.34 | 1.12 | 1.12 | 1.65 | -1.14 | 1.05 | -1.1 |
| specification of segmental identity, abdomen | 5 | 1.18 | 1.16 | 1.14 | 1.3 | -1.04 | 1.14 | -1.03 |
| compartment pattern specification | 4 | -1.06 | -1.16 | 1.12 | 1.16 | 1.23 | -1.31 | -1.4 |
| posterior compartment specification | 1 | -1.02 | -1.34 | -1.22 | -1.07 | -1.24 | -1.21 | -1.17 |
| pattern specification process | 406 | 1.19 | 1.05 | 1.18 | 1.01 | 1.22 | 1.26 | 1.25 |
| germ-band shortening | 18 | 1.2 | 1.11 | 1.38 | 1.07 | 1.46 | 1.23 | 1.55 |
| dorsal closure | 88 | 1.12 | 1.1 | 1.41 | 1.05 | 1.37 | 1.28 | 1.31 |
| initiation of dorsal closure | 17 | 1.16 | 1.17 | 1.59 | 1.07 | 1.56 | 1.2 | 1.36 |
| dorsal closure, leading edge cell fate determination | 3 | 1.0 | 1.08 | 1.89 | 1.0 | 1.21 | -1.44 | -1.04 |
| dorsal closure, elongation of leading edge cells | 12 | 1.22 | 1.22 | 1.4 | 1.12 | 1.59 | 1.38 | 1.41 |
| dorsal closure, spreading of leading edge cells | 4 | 1.03 | 1.07 | 2.59 | -1.02 | 1.79 | 1.38 | 1.66 |
| suture of dorsal opening | 3 | -1.66 | 1.12 | 1.92 | -1.03 | 1.4 | -1.35 | 1.31 |
| organ morphogenesis | 472 | 1.18 | 1.09 | 1.26 | 1.04 | 1.27 | 1.27 | 1.24 |
| tissue development | 483 | 1.17 | 1.03 | 1.19 | 1.01 | 1.23 | 1.27 | 1.18 |
| ectoderm development | 21 | 1.37 | -1.09 | 1.05 | 1.01 | 1.24 | 1.01 | 1.03 |
| nervous system development | 1192 | 1.14 | 1.06 | 1.21 | 1.07 | 1.22 | 1.19 | 1.13 |
| neuroblast fate determination | 25 | 1.13 | 1.09 | 1.19 | 1.07 | 1.23 | -1.0 | 1.07 |
| neurological system process | 607 | 1.09 | 1.03 | 1.1 | 1.0 | 1.09 | 1.15 | 1.07 |
| ganglion mother cell fate determination | 8 | 1.18 | 1.1 | 1.09 | 1.08 | 1.22 | 1.05 | 1.11 |
| glial cell fate determination | 3 | 1.44 | 1.3 | 1.68 | 1.12 | 1.63 | 1.5 | 1.59 |
| neuroblast proliferation | 40 | -1.13 | 1.17 | 1.36 | 1.16 | 1.4 | 1.07 | 1.11 |
| negative regulation of neuroblast proliferation | 7 | -1.32 | 1.13 | 1.47 | 1.06 | 1.79 | 1.22 | 1.08 |
| axonogenesis | 233 | 1.11 | 1.08 | 1.25 | 1.0 | 1.27 | 1.27 | 1.31 |
| axon guidance | 181 | 1.1 | 1.08 | 1.23 | 1.01 | 1.27 | 1.29 | 1.31 |
| axon target recognition | 15 | -1.0 | 1.14 | 1.31 | 1.09 | 1.01 | -1.17 | 1.08 |
| axonal fasciculation | 14 | -1.38 | -1.31 | -1.09 | -1.08 | 1.2 | 1.11 | -1.02 |
| axonal defasciculation | 11 | -1.03 | -1.0 | 1.06 | -1.05 | 1.13 | 1.11 | 1.26 |
| defasciculation of motor neuron axon | 7 | 1.05 | -1.01 | 1.02 | 1.01 | 1.05 | 1.08 | 1.18 |
| synapse assembly | 33 | 1.25 | 1.16 | 1.26 | 1.01 | 1.24 | 1.48 | 1.24 |
| central nervous system development | 184 | 1.13 | 1.07 | 1.29 | 1.04 | 1.2 | 1.15 | 1.15 |
| ventral midline development | 6 | -1.04 | -1.05 | 1.01 | 1.07 | 1.13 | -1.04 | 1.11 |
| ventral cord development | 31 | 1.44 | 1.02 | 1.19 | -1.02 | 1.16 | 1.12 | 1.17 |
| brain development | 99 | 1.09 | 1.08 | 1.33 | 1.05 | 1.24 | 1.12 | 1.14 |
| chemosensory behavior | 89 | 1.01 | 1.02 | 1.17 | -1.01 | 1.16 | 1.11 | 1.15 |
| stomatogastric nervous system development | 3 | 1.52 | 1.02 | 1.2 | 1.12 | 1.56 | 1.54 | 1.47 |
| peripheral nervous system development | 83 | 1.1 | 1.04 | 1.18 | 1.03 | 1.32 | 1.2 | 1.23 |
| sensory organ development | 366 | 1.13 | 1.09 | 1.25 | 1.06 | 1.24 | 1.18 | 1.19 |
| open tracheal system development | 179 | 1.41 | 1.03 | 1.26 | -1.02 | 1.26 | 1.52 | 1.28 |
| epithelial cell fate determination, open tracheal system | 5 | 1.83 | 1.1 | 1.52 | -1.02 | 1.42 | 1.21 | 1.34 |
| tracheal outgrowth, open tracheal system | 24 | 1.41 | 1.06 | 1.45 | 1.03 | 1.41 | 1.42 | 1.26 |
| epithelial cell migration, open tracheal system | 28 | 1.66 | -1.03 | 1.29 | -1.07 | 1.21 | 1.38 | 1.11 |
| primary branching, open tracheal system | 14 | 1.41 | 1.06 | 1.72 | 1.04 | 1.34 | 1.6 | 1.2 |
| secondary branching, open tracheal system | 4 | -1.26 | 1.18 | 1.54 | -1.02 | 1.25 | 1.45 | 1.01 |
| terminal branching, open tracheal system | 10 | 1.25 | 1.17 | 1.56 | 1.07 | 1.46 | 1.37 | 1.13 |
| salivary gland development | 141 | 1.08 | 1.0 | 1.15 | 1.05 | 1.27 | 1.18 | 1.17 |
| salivary gland boundary specification | 12 | 1.24 | 1.02 | 1.3 | 1.14 | 1.55 | 1.11 | 1.19 |
| salivary gland morphogenesis | 122 | 1.06 | 1.0 | 1.13 | 1.04 | 1.25 | 1.23 | 1.18 |
| larval salivary gland morphogenesis | 6 | -1.03 | 1.12 | 1.25 | 1.04 | 1.36 | 1.52 | 1.57 |
| oenocyte development | 7 | 1.11 | 1.13 | 1.16 | 1.11 | 1.2 | 1.21 | 1.08 |
| ectodermal digestive tract development | 5 | 1.46 | 1.11 | -1.0 | 1.02 | 1.62 | 1.8 | 1.33 |
| foregut morphogenesis | 10 | -1.4 | -1.11 | -1.17 | -1.1 | 1.29 | -1.3 | 1.37 |
| hindgut morphogenesis | 56 | 1.24 | -1.1 | 1.09 | -1.03 | 1.35 | 1.37 | 1.13 |
| Malpighian tubule morphogenesis | 44 | 1.29 | -1.1 | 1.07 | -1.04 | 1.27 | 1.38 | 1.09 |
| imaginal disc development | 435 | 1.23 | 1.04 | 1.19 | 1.01 | 1.23 | 1.3 | 1.24 |
| determination of imaginal disc primordium | 10 | 1.07 | 1.03 | 1.07 | -1.02 | 1.18 | 1.04 | 1.37 |
| imaginal disc growth | 17 | 1.45 | 1.18 | 1.4 | 1.14 | 1.42 | 1.3 | 1.32 |
| imaginal disc pattern formation | 93 | 1.23 | 1.0 | 1.11 | 1.04 | 1.22 | 1.28 | 1.18 |
| anterior/posterior pattern specification, imaginal disc | 20 | 1.23 | -1.26 | 1.02 | -1.03 | 1.24 | 1.42 | -1.03 |
| proximal/distal pattern formation, imaginal disc | 21 | 1.34 | -1.0 | 1.11 | 1.1 | 1.1 | -1.01 | 1.11 |
| dorsal/ventral pattern formation, imaginal disc | 46 | 1.31 | 1.01 | 1.09 | -1.0 | 1.25 | 1.33 | 1.29 |
| dorsal/ventral lineage restriction, imaginal disc | 7 | 1.04 | 1.11 | 1.08 | 1.07 | 1.41 | 1.38 | 1.52 |
| eye-antennal disc morphogenesis | 37 | 1.29 | 1.08 | 1.27 | 1.08 | 1.29 | 1.31 | 1.24 |
| progression of morphogenetic furrow involved in compound eye morphogenesis | 8 | 1.62 | -1.01 | 1.33 | 1.05 | 1.35 | 1.15 | 1.12 |
| R8 cell fate commitment | 14 | 1.18 | 1.05 | 1.18 | 1.13 | 1.28 | -1.01 | 1.04 |
| R1/R6 cell fate commitment | 2 | 1.08 | 1.03 | -1.12 | -1.19 | -1.15 | -1.01 | 1.1 |
| R3/R4 cell fate commitment | 14 | 1.45 | 1.1 | 1.12 | 1.04 | 1.4 | 1.31 | 1.24 |
| R7 cell fate commitment | 20 | 1.3 | 1.11 | 1.31 | 1.06 | 1.32 | 1.34 | 1.27 |
| regulation of rhodopsin gene expression | 2 | 1.63 | 1.26 | 1.37 | 1.04 | 1.38 | 1.48 | 1.42 |
| antennal development | 21 | -1.03 | 1.02 | 1.12 | 1.04 | 1.07 | 1.26 | 1.08 |
| wing disc morphogenesis | 207 | 1.31 | 1.07 | 1.29 | 1.02 | 1.28 | 1.37 | 1.34 |
| wing disc proximal/distal pattern formation | 6 | 1.82 | -1.07 | 1.3 | 1.34 | 1.32 | 1.02 | -1.06 |
| imaginal disc-derived wing vein specification | 33 | 1.21 | 1.03 | 1.14 | 1.0 | 1.19 | 1.17 | 1.38 |
| apposition of dorsal and ventral imaginal disc-derived wing surfaces | 17 | 1.92 | 1.04 | 1.15 | -1.07 | 1.24 | 1.93 | 1.28 |
| imaginal disc-derived wing morphogenesis | 203 | 1.31 | 1.07 | 1.28 | 1.02 | 1.28 | 1.38 | 1.34 |
| notum development | 8 | 1.04 | 1.0 | 1.09 | 1.1 | 1.29 | -1.05 | 1.15 |
| leg disc morphogenesis | 54 | -1.03 | 1.07 | 1.1 | 1.02 | 1.38 | 1.23 | 1.22 |
| leg disc proximal/distal pattern formation | 16 | 1.45 | 1.01 | 1.11 | 1.07 | 1.06 | 1.05 | 1.22 |
| imaginal disc-derived leg morphogenesis | 53 | -1.02 | 1.08 | 1.1 | 1.01 | 1.37 | 1.25 | 1.26 |
| haltere disc morphogenesis | 2 | 2.21 | -1.24 | 1.11 | 1.19 | -1.26 | -1.2 | -1.1 |
| haltere development | 9 | 1.36 | -1.03 | -1.02 | 1.14 | 1.16 | -1.19 | 1.04 |
| genital disc morphogenesis | 17 | 1.4 | 1.1 | 1.29 | 1.07 | 1.2 | 1.04 | 1.22 |
| imaginal disc-derived genitalia development | 22 | 1.56 | 1.05 | 1.18 | 1.02 | 1.13 | 1.11 | 1.24 |
| imaginal disc-derived male genitalia development | 20 | 1.63 | 1.05 | 1.2 | 1.02 | 1.16 | 1.13 | 1.28 |
| imaginal disc-derived female genitalia development | 6 | 1.5 | 1.12 | 1.14 | 1.04 | 1.07 | 1.14 | 1.13 |
| analia development | 6 | -1.15 | -1.64 | -1.26 | 1.2 | 1.4 | -1.24 | -1.79 |
| histoblast morphogenesis | 8 | -1.19 | 1.09 | 1.37 | 1.16 | 1.65 | -1.2 | 1.21 |
| maintenance of imaginal histoblast diploidy | 1 | -10.7 | -1.04 | 2.18 | 1.33 | 2.43 | -8.45 | -1.16 |
| endoderm development | 8 | 1.22 | -1.04 | 1.07 | 1.01 | 1.2 | 1.25 | 1.45 |
| endodermal cell fate determination | 1 | 8.12 | -1.58 | 1.44 | -1.54 | 1.01 | 1.69 | 1.21 |
| midgut development | 31 | -1.29 | -1.1 | 1.1 | -1.15 | -1.0 | 1.01 | 1.05 |
| anterior midgut development | 3 | -1.08 | 1.25 | 1.25 | -1.4 | 1.12 | 1.13 | 1.58 |
| posterior midgut development | 1 | 1.06 | -1.06 | -1.11 | -1.03 | -1.09 | -1.08 | -1.11 |
| ectoderm and mesoderm interaction | 2 | 1.28 | 1.06 | 1.06 | 1.18 | 1.33 | 1.49 | 1.08 |
| mesodermal cell fate determination | 5 | 1.66 | 1.11 | 1.13 | -1.01 | 1.25 | 1.53 | 1.29 |
| mesodermal cell fate specification | 11 | 1.38 | -1.02 | -1.0 | -1.04 | 1.01 | 1.33 | -1.02 |
| digestive tract mesoderm development | 3 | 1.05 | 1.02 | 1.08 | 1.02 | 1.19 | -1.09 | -1.12 |
| fat body development | 3 | 1.03 | 1.03 | -1.06 | -1.03 | -1.1 | -1.05 | 1.49 |
| gonadal mesoderm development | 8 | 1.06 | -1.02 | 1.26 | 1.06 | 1.16 | 1.0 | -1.13 |
| heart development | 70 | 1.28 | -1.02 | 1.19 | -1.07 | 1.17 | 1.3 | 1.17 |
| larval heart development | 2 | 1.04 | -1.11 | 1.46 | -1.3 | -1.07 | -1.65 | 1.78 |
| mesoderm migration involved in gastrulation | 10 | 1.62 | -1.05 | 1.42 | -1.16 | 1.22 | 2.32 | 1.08 |
| cardioblast cell fate determination | 4 | 1.32 | 1.0 | 1.08 | -1.04 | -1.05 | -1.0 | 1.04 |
| lymph gland development | 22 | 1.29 | 1.04 | 1.21 | 1.05 | 1.2 | 1.21 | 1.38 |
| hemocyte development | 5 | 1.5 | 1.11 | 1.52 | 1.05 | 1.28 | 1.12 | 1.43 |
| muscle organ development | 163 | 1.31 | 1.06 | 1.2 | 1.02 | 1.21 | 1.31 | 1.18 |
| myoblast cell fate determination | 1 | -1.07 | -1.09 | 1.09 | -1.1 | 1.09 | -1.05 | 1.02 |
| skeletal muscle tissue development | 79 | 1.17 | 1.09 | 1.25 | 1.03 | 1.19 | 1.34 | 1.16 |
| myoblast fusion | 27 | 1.01 | 1.06 | 1.27 | -1.02 | 1.21 | 1.27 | 1.14 |
| muscle cell fate determination | 3 | 1.16 | 1.17 | 1.3 | 1.2 | 1.33 | -1.2 | 1.28 |
| visceral muscle development | 11 | 1.37 | -1.08 | 1.14 | -1.13 | 1.1 | 1.25 | 1.21 |
| larval visceral muscle development | 5 | 1.28 | -1.03 | 1.19 | -1.12 | 1.18 | 1.12 | 1.5 |
| somatic muscle development | 32 | 1.35 | 1.09 | 1.29 | 1.02 | 1.2 | 1.12 | 1.09 |
| larval somatic muscle development | 8 | 1.66 | 1.08 | 1.35 | 1.03 | 1.17 | 1.3 | 1.22 |
| adult somatic muscle development | 10 | 1.25 | 1.07 | 1.59 | 1.07 | 1.35 | 1.16 | 1.08 |
| neuromuscular junction development | 47 | 1.2 | 1.17 | 1.29 | 1.08 | 1.21 | 1.33 | 1.23 |
| sex determination | 25 | 1.28 | 1.14 | 1.27 | 1.1 | 1.2 | 1.19 | 1.13 |
| primary sex determination | 17 | 1.27 | 1.11 | 1.28 | 1.06 | 1.22 | 1.26 | 1.15 |
| primary sex determination, soma | 13 | 1.13 | 1.12 | 1.28 | 1.07 | 1.19 | 1.09 | 1.1 |
| sex determination, establishment of X:A ratio | 5 | -1.07 | 1.05 | 1.18 | 1.02 | 1.03 | -1.1 | -1.09 |
| sex determination, primary response to X:A ratio | 5 | 1.36 | 1.19 | 1.41 | 1.08 | 1.34 | 1.16 | 1.31 |
| primary sex determination, germ-line | 4 | 1.82 | 1.02 | 1.1 | 1.07 | 1.07 | 1.73 | 1.13 |
| sex differentiation | 82 | 1.2 | -1.07 | -1.04 | -1.09 | 1.12 | 1.09 | 1.08 |
| dosage compensation | 11 | 1.22 | 1.11 | 1.25 | -1.03 | 1.17 | 1.34 | 1.47 |
| metamorphosis | 374 | 1.17 | 1.04 | 1.18 | 1.01 | 1.26 | 1.25 | 1.24 |
| regulation of ecdysteroid metabolic process | 1 | -1.55 | 1.2 | 1.21 | -1.19 | -1.12 | 1.05 | 1.03 |
| regulation of ecdysteroid secretion | 1 | -1.14 | 1.41 | 1.3 | 1.24 | 1.17 | 1.85 | 1.43 |
| regulation of juvenile hormone metabolic process | 2 | -8.29 | -1.83 | 1.21 | 1.4 | 1.08 | -7.35 | -3.84 |
| regulation of juvenile hormone biosynthetic process | 2 | -8.29 | -1.83 | 1.21 | 1.4 | 1.08 | -7.35 | -3.84 |
| histolysis | 79 | -1.03 | 1.02 | 1.13 | 1.07 | 1.24 | 1.12 | 1.1 |
| imaginal disc morphogenesis | 285 | 1.23 | 1.07 | 1.25 | 1.02 | 1.3 | 1.3 | 1.29 |
| imaginal disc eversion | 8 | -1.35 | 1.14 | 1.18 | 1.03 | 1.2 | 1.09 | 1.17 |
| eclosion | 15 | 1.14 | 1.03 | 1.09 | -1.05 | -1.02 | 1.17 | 1.27 |
| regulation of eclosion | 5 | 1.16 | 1.13 | 1.1 | 1.07 | 1.05 | -1.04 | -1.04 |
| regulation of chitin-based cuticle tanning | 13 | -1.19 | -1.03 | -1.1 | -1.01 | 1.02 | -1.1 | 1.19 |
| aging | 114 | 1.25 | 1.12 | 1.11 | -1.02 | 1.16 | 1.37 | 1.25 |
| age-dependent general metabolic decline | 1 | -1.1 | 1.09 | 1.19 | 1.01 | 1.01 | 1.19 | 1.25 |
| autophagic cell death | 70 | 1.04 | -1.0 | 1.1 | 1.06 | 1.23 | 1.21 | 1.11 |
| response to nutrient | 14 | -1.16 | 1.15 | 1.37 | 1.05 | 1.35 | 1.65 | 1.19 |
| digestion | 3 | -4.6 | 1.02 | -2.88 | -1.01 | -3.41 | -5.92 | -1.12 |
| excretion | 1 | 1.59 | -1.18 | -1.24 | -1.07 | -1.14 | 4.1 | -1.27 |
| body fluid secretion | 5 | -1.82 | -1.14 | 1.09 | 1.02 | 1.2 | -2.0 | -1.34 |
| storage protein import into fat body | 1 | -1.43 | -1.07 | 1.81 | 1.21 | 1.62 | -1.56 | -1.76 |
| molting cycle, chitin-based cuticle | 46 | 1.19 | -1.01 | 1.03 | -1.09 | -1.03 | 1.24 | 1.14 |
| protein-based cuticle development | 13 | 1.28 | -1.16 | -1.04 | -1.21 | -1.09 | 1.14 | 1.17 |
| chitin-based cuticle tanning | 9 | 1.05 | -1.17 | -1.07 | -1.06 | -1.1 | -1.03 | 1.38 |
| puparial adhesion | 11 | -1.06 | 1.01 | -1.06 | -1.01 | -1.12 | -1.06 | -1.12 |
| hemostasis | 5 | 1.95 | -1.51 | -1.67 | -1.6 | -1.41 | 1.79 | -1.02 |
| sensory perception | 298 | 1.08 | -1.01 | -1.0 | -1.02 | -1.02 | 1.03 | -1.03 |
| visual perception | 40 | 1.13 | -1.01 | 1.16 | 1.08 | 1.1 | 1.09 | 1.01 |
| phototransduction | 43 | 1.04 | -1.12 | -1.06 | 1.05 | 1.04 | -1.05 | -1.09 |
| phototransduction, visible light | 19 | 1.05 | -1.08 | -1.05 | 1.11 | 1.04 | -1.1 | -1.12 |
| phototransduction, UV | 5 | -1.46 | -1.43 | -1.51 | 1.22 | -1.06 | -1.54 | -1.62 |
| sensory perception of sound | 21 | -1.01 | 1.1 | 1.22 | 1.04 | 1.15 | 1.25 | 1.02 |
| sensory perception of chemical stimulus | 235 | 1.08 | -1.02 | -1.05 | -1.04 | -1.05 | -1.0 | -1.04 |
| sensory perception of taste | 68 | -1.0 | 1.03 | 1.01 | 1.0 | 1.03 | -1.02 | -1.02 |
| detection of chemical stimulus involved in sensory perception of taste | 5 | 1.32 | -1.06 | -1.09 | -1.07 | 1.79 | 1.58 | 1.13 |
| sensory perception of smell | 82 | 1.05 | 1.01 | 1.0 | 1.0 | -1.03 | -1.02 | -1.01 |
| behavior | 399 | 1.11 | -1.01 | 1.12 | -1.04 | 1.1 | 1.22 | 1.16 |
| learning or memory | 104 | 1.16 | 1.04 | 1.18 | -1.01 | 1.23 | 1.38 | 1.22 |
| learning | 62 | 1.11 | 1.07 | 1.18 | 1.02 | 1.21 | 1.32 | 1.22 |
| memory | 61 | 1.21 | 1.03 | 1.22 | -1.03 | 1.24 | 1.42 | 1.28 |
| short-term memory | 12 | 1.22 | 1.21 | 1.62 | -1.01 | 1.53 | 1.26 | 1.48 |
| anesthesia-resistant memory | 8 | 1.01 | -1.16 | -1.04 | -1.21 | -1.14 | 1.22 | 1.39 |
| long-term memory | 38 | 1.27 | 1.07 | 1.27 | 1.02 | 1.25 | 1.51 | 1.25 |
| mating behavior | 79 | 1.01 | -1.1 | -1.02 | -1.04 | -1.07 | 1.02 | 1.1 |
| mating | 90 | 1.04 | -1.1 | -1.03 | -1.06 | -1.06 | 1.08 | 1.06 |
| courtship behavior | 53 | 1.01 | -1.16 | -1.02 | -1.12 | -1.1 | 1.01 | 1.15 |
| copulation | 23 | 1.23 | -1.11 | -1.09 | -1.17 | -1.08 | 1.34 | -1.08 |
| negative regulation of female receptivity | 8 | -1.24 | 1.06 | 1.02 | 1.03 | 1.16 | -1.13 | -1.08 |
| rhythmic behavior | 47 | -1.03 | -1.07 | 1.06 | -1.22 | -1.14 | 1.21 | 1.12 |
| circadian rhythm | 55 | 1.08 | -1.07 | 1.04 | -1.2 | -1.1 | 1.29 | 1.14 |
| ultradian rhythm | 1 | -1.1 | 1.09 | 1.19 | 1.01 | 1.01 | 1.19 | 1.25 |
| grooming behavior | 2 | 2.8 | 1.39 | 1.39 | -1.27 | -1.08 | 1.53 | 1.56 |
| locomotory behavior | 119 | 1.1 | 1.06 | 1.22 | -1.07 | 1.12 | 1.36 | 1.15 |
| larval behavior | 32 | -1.13 | 1.1 | 1.17 | -1.13 | -1.02 | -1.04 | 1.19 |
| adult walking behavior | 9 | -1.95 | 1.22 | 1.15 | 1.1 | 1.23 | -1.39 | 1.15 |
| flight behavior | 18 | 1.38 | -1.05 | 1.24 | -1.05 | 1.31 | 1.48 | 1.11 |
| jump response | 7 | 1.63 | -1.18 | 1.1 | -1.09 | 1.06 | 1.45 | -1.03 |
| feeding behavior | 27 | 1.06 | -1.12 | 1.12 | -1.22 | -1.11 | -1.09 | 1.36 |
| visual behavior | 19 | 1.39 | 1.02 | 1.17 | 1.04 | 1.14 | 1.16 | 1.23 |
| chemosensory jump behavior | 1 | -1.0 | 1.03 | 1.05 | 1.01 | 1.02 | 1.05 | 1.11 |
| proboscis extension reflex | 5 | -1.0 | -1.02 | 1.2 | 1.04 | 1.13 | 1.24 | 1.12 |
| mechanosensory behavior | 6 | -1.75 | 1.04 | 1.58 | 1.06 | 1.09 | 1.12 | -1.04 |
| structural constituent of chitin-based larval cuticle | 39 | -1.05 | -1.05 | -1.13 | -1.07 | -1.2 | -1.1 | -1.03 |
| structural constituent of pupal chitin-based cuticle | 7 | -1.04 | -1.01 | 1.04 | 1.0 | -1.03 | -1.08 | -1.07 |
| structural constituent of adult chitin-based cuticle | 4 | 1.11 | 1.06 | 1.05 | -1.05 | 1.06 | 1.05 | -1.03 |
| beta-catenin binding | 9 | 1.09 | 1.18 | 1.31 | 1.15 | 1.46 | 1.17 | 1.28 |
| calcium-dependent cell-cell adhesion | 18 | 1.55 | -1.07 | -1.04 | -1.12 | 1.13 | 1.76 | 1.45 |
| blood circulation | 3 | -1.25 | 1.41 | -1.37 | -1.01 | 1.12 | 1.05 | 1.18 |
| regulation of heart contraction | 13 | -1.49 | 1.0 | 1.16 | -1.05 | -1.04 | -1.08 | -1.3 |
| microtubule binding | 63 | 1.08 | 1.04 | 1.33 | -1.0 | 1.35 | 1.38 | 1.26 |
| pattern recognition receptor activity | 6 | 6.25 | -1.32 | -1.34 | -1.44 | -1.26 | 3.38 | 1.39 |
| G-protein coupled photoreceptor activity | 8 | -1.07 | -1.12 | 1.03 | -1.04 | -1.29 | 1.03 | -1.14 |
| synaptic vesicle | 56 | 1.07 | 1.09 | 1.23 | 1.07 | 1.27 | 1.16 | 1.16 |
| protein C-terminus binding | 1 | -1.42 | 1.29 | 1.09 | 1.21 | 1.26 | -1.02 | 1.15 |
| transcription elongation factor complex | 27 | 1.25 | 1.16 | 1.35 | 1.13 | 1.31 | 1.3 | 1.17 |
| positive transcription elongation factor complex b | 3 | 1.59 | 1.11 | 1.57 | 1.18 | 1.65 | 1.7 | 1.24 |
| diazepam binding | 5 | -4.23 | -1.51 | -3.23 | 1.14 | 1.44 | -4.78 | -1.66 |
| ATP-dependent helicase activity | 76 | 1.18 | 1.21 | 1.34 | 1.17 | 1.3 | 1.31 | 1.19 |
| monocarboxylic acid transmembrane transporter activity | 16 | -2.06 | 1.03 | -1.25 | 1.14 | 1.09 | -1.24 | -1.32 |
| pentraxin receptor activity | 1 | 1.18 | 1.16 | 1.11 | 1.22 | 1.12 | 1.2 | 1.16 |
| neuronal pentraxin receptor activity | 1 | 1.18 | 1.16 | 1.11 | 1.22 | 1.12 | 1.2 | 1.16 |
| eclosion hormone activity | 1 | -1.16 | 1.0 | -1.13 | -1.07 | -1.21 | -1.36 | -1.2 |
| tRNA processing | 33 | 1.3 | 1.13 | 1.13 | 1.23 | 1.16 | 1.1 | -1.08 |
| diuretic hormone receptor activity | 3 | 1.32 | -1.32 | 1.22 | -1.4 | -1.1 | -1.32 | 2.54 |
| cell recognition | 57 | 1.01 | -1.01 | 1.18 | 1.01 | 1.28 | 1.17 | 1.15 |
| neuron recognition | 56 | 1.01 | -1.01 | 1.18 | 1.01 | 1.29 | 1.18 | 1.16 |
| synaptic target recognition | 8 | 1.28 | -1.15 | 1.12 | -1.06 | 1.36 | 1.36 | 1.15 |
| intracellular ferritin complex | 3 | -1.78 | 1.04 | 1.19 | 1.02 | -1.06 | -2.25 | -1.21 |
| adult behavior | 100 | 1.26 | -1.03 | 1.14 | -1.07 | 1.14 | 1.22 | 1.28 |
| motor axon guidance | 40 | 1.33 | 1.03 | 1.17 | -1.02 | 1.25 | 1.44 | 1.49 |
| axon guidance receptor activity | 4 | -1.18 | -1.02 | 1.16 | -1.02 | 1.15 | -1.24 | 1.14 |
| enzyme activator activity | 85 | 1.11 | 1.14 | 1.32 | 1.07 | 1.24 | 1.11 | 1.2 |
| calcium sensitive guanylate cyclase activator activity | 1 | 4.81 | -1.03 | -1.02 | 1.21 | 1.81 | 1.05 | 1.23 |
| male courtship behavior | 38 | -1.12 | -1.17 | 1.06 | -1.08 | -1.08 | -1.08 | 1.09 |
| sensory organ boundary specification | 21 | 1.12 | 1.05 | 1.15 | 1.04 | 1.26 | 1.14 | 1.19 |
| mitochondrial fusion | 4 | -1.03 | 1.3 | 1.41 | 1.13 | 1.32 | 1.68 | 1.08 |
| cyclin catabolic process | 6 | -1.14 | 1.19 | 1.17 | 1.12 | 1.29 | -1.06 | 1.05 |
| ocellus pigment biosynthetic process | 18 | 1.35 | 1.13 | 1.12 | 1.06 | 1.06 | 1.08 | 1.16 |
| ocellus development | 3 | 1.06 | 1.06 | -1.0 | 1.09 | -1.06 | -1.05 | -1.05 |
| eye pigment granule organization | 10 | 1.1 | 1.2 | 1.42 | 1.14 | 1.25 | 1.18 | 1.2 |
| ARF GTPase activator activity | 9 | -1.07 | 1.11 | 1.17 | 1.02 | 1.17 | -1.02 | 1.45 |
| chitin binding | 82 | -2.26 | -1.76 | -2.0 | -1.6 | -1.65 | -2.02 | 1.28 |
| eclosion rhythm | 10 | 1.18 | -1.0 | 1.16 | -1.06 | 1.02 | 1.29 | 1.48 |
| Toll signaling pathway | 33 | 2.4 | -1.26 | -1.12 | -1.26 | -1.03 | 2.14 | 1.22 |
| regulation of actin polymerization or depolymerization | 20 | -1.48 | 1.08 | 1.45 | 1.05 | 1.38 | -1.21 | 1.18 |
| establishment of blood-nerve barrier | 3 | 1.7 | 1.01 | 1.41 | -1.21 | 1.32 | 4.52 | 2.14 |
| glutamate receptor activity | 40 | 1.1 | 1.01 | 1.02 | -1.03 | -1.05 | -1.01 | -1.02 |
| extracellular-glutamate-gated chloride channel activity | 1 | 1.11 | 1.03 | -1.01 | -1.03 | 1.01 | 1.02 | 1.03 |
| dorsal/ventral axis specification, ovarian follicular epithelium | 8 | 1.49 | 1.13 | 1.18 | 1.13 | 1.25 | 1.48 | 1.25 |
| maternal determination of dorsal/ventral axis, ovarian follicular epithelium, germ-line encoded | 1 | 8.32 | -1.09 | 1.22 | 1.02 | 1.27 | 3.95 | 1.61 |
| maternal determination of dorsal/ventral axis, ovarian follicular epithelium, soma encoded | 1 | 2.54 | -1.11 | -1.09 | 1.14 | 1.59 | 3.23 | 1.89 |
| ornithine decarboxylase inhibitor activity | 1 | 1.05 | 1.03 | -1.12 | 1.06 | -1.03 | -1.01 | 1.05 |
| guanylate cyclase complex, soluble | 5 | -1.02 | -1.02 | -1.05 | -1.02 | -1.07 | -1.02 | -1.11 |
| voltage-gated potassium channel complex | 18 | 1.03 | 1.03 | 1.1 | 1.02 | 1.05 | -1.06 | -1.0 |
| mesodermal cell migration | 6 | 1.63 | -1.09 | 1.53 | -1.13 | 1.3 | 1.85 | -1.09 |
| translation termination factor activity | 7 | 1.27 | 1.16 | 1.25 | 1.13 | 1.18 | 1.49 | 1.09 |
| N-acetyltransferase activity | 43 | -1.1 | 1.17 | 1.01 | 1.17 | 1.18 | -1.21 | 1.02 |
| phosphoric diester hydrolase activity | 30 | -1.19 | -1.12 | -1.13 | 1.09 | -1.0 | -1.15 | -1.05 |
| growth factor activity | 27 | 1.05 | 1.01 | 1.04 | -1.13 | -1.02 | 1.19 | 1.11 |
| imaginal disc growth factor receptor binding | 5 | 4.12 | -4.42 | -5.25 | -6.0 | -2.16 | 10.45 | 2.09 |
| light-activated voltage-gated calcium channel activity | 2 | -1.34 | -1.15 | -1.19 | 1.14 | -1.2 | -1.27 | -1.19 |
| light-activated voltage-gated calcium channel complex | 1 | -1.32 | -1.06 | -1.2 | 1.5 | -1.29 | -1.26 | -1.29 |
| axon cargo transport | 16 | 1.1 | 1.21 | 1.27 | 1.13 | 1.19 | 1.36 | 1.14 |
| anterograde axon cargo transport | 4 | 1.14 | 1.18 | 1.17 | 1.17 | 1.3 | 1.25 | 1.03 |
| retrograde axon cargo transport | 2 | -1.28 | 1.31 | 1.1 | 1.11 | 1.09 | 1.07 | 1.09 |
| spectrin | 3 | -2.12 | 1.38 | 2.51 | 1.03 | 1.66 | 1.56 | 1.52 |
| cytoskeletal protein binding | 213 | 1.08 | 1.03 | 1.32 | -1.0 | 1.29 | 1.21 | 1.16 |
| cytoskeletal adaptor activity | 3 | -1.31 | 1.01 | 1.0 | 1.13 | 1.08 | -1.15 | -1.23 |
| DNA-dependent ATPase activity | 34 | 1.07 | 1.15 | 1.32 | 1.15 | 1.32 | 1.15 | 1.17 |
| juvenile hormone epoxide hydrolase activity | 3 | -2.11 | 1.3 | -1.14 | -1.06 | -1.44 | -4.35 | 1.38 |
| 5S rRNA binding | 2 | 1.26 | 1.08 | 1.22 | 1.14 | 1.15 | 1.1 | -1.0 |
| 5S rRNA primary transcript binding | 1 | 1.17 | 1.17 | 1.25 | 1.35 | 1.14 | -1.19 | -1.07 |
| decapentaplegic signaling pathway | 11 | 1.19 | 1.18 | 1.63 | 1.11 | 1.61 | 1.49 | 1.44 |
| oocyte microtubule cytoskeleton polarization | 19 | 1.07 | 1.18 | 1.47 | 1.08 | 1.29 | 1.46 | 1.27 |
| asymmetric protein localization | 36 | -1.02 | 1.11 | 1.3 | 1.09 | 1.44 | 1.21 | 1.16 |
| alcohol dehydrogenase (NADP+) activity | 7 | 1.25 | 1.15 | -1.37 | -1.18 | -1.21 | 1.14 | 1.22 |
| N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity | 1 | -2.23 | -1.22 | -3.38 | -1.38 | 1.35 | 1.93 | 6.67 |
| alpha-methylacyl-CoA racemase activity | 1 | 4.0 | -1.32 | -1.02 | 1.57 | 2.33 | 1.69 | -1.27 |
| peptide-methionine-(S)-S-oxide reductase activity | 2 | -1.8 | -2.02 | -4.93 | 1.56 | -1.77 | -2.69 | -2.87 |
| sphinganine-1-phosphate aldolase activity | 1 | -4.18 | -1.48 | 1.63 | 1.57 | 1.8 | -2.56 | -3.36 |
| ceramide glucosyltransferase activity | 1 | -2.39 | 1.12 | 1.45 | -1.07 | -1.2 | -1.93 | -1.3 |
| ubiquinol-cytochrome-c reductase activity | 10 | -1.05 | 1.06 | 1.14 | 1.09 | 1.02 | 1.34 | -1.41 |
| 4-alpha-hydroxytetrahydrobiopterin dehydratase activity | 1 | 7.73 | -1.1 | 1.09 | -1.29 | -1.0 | 2.46 | 1.16 |
| primary amine oxidase activity | 1 | -1.01 | 1.1 | 1.17 | 1.35 | 1.05 | -1.29 | -1.96 |
| transcription factor binding | 100 | 1.32 | 1.1 | 1.22 | 1.06 | 1.24 | 1.32 | 1.24 |
| translation factor activity, nucleic acid binding | 74 | 1.34 | 1.17 | 1.26 | 1.09 | 1.19 | 1.38 | 1.12 |
| NADH dehydrogenase (ubiquinone) activity | 30 | 1.12 | 1.03 | 1.27 | 1.07 | 1.1 | 1.73 | -1.3 |
| protein tyrosine/serine/threonine phosphatase activity | 20 | 1.45 | 1.13 | 1.29 | 1.1 | 1.37 | 1.36 | 1.21 |
| nuclear localization sequence binding | 1 | -1.11 | 1.3 | 1.01 | 1.22 | 1.27 | 1.55 | 1.52 |
| cAMP response element binding protein binding | 1 | 2.75 | 1.33 | 1.9 | 1.11 | 2.22 | 3.34 | 2.0 |
| oxysterol binding | 3 | -2.13 | 1.47 | 1.84 | 1.14 | 1.36 | 1.14 | 1.32 |
| drug binding | 19 | -1.19 | -1.07 | -1.35 | 1.08 | 1.12 | -1.56 | -1.08 |
| phenylalkylamine binding | 5 | 1.32 | 1.03 | -1.03 | 1.02 | -1.05 | -1.01 | -1.01 |
| sulfotransferase activity | 17 | 1.29 | 1.02 | 1.09 | -1.04 | 1.07 | -1.07 | 1.2 |
| negative regulation of transcription elongation, DNA-dependent | 4 | 1.21 | 1.13 | 1.22 | 1.16 | 1.17 | 1.14 | 1.08 |
| biological_process | 8429 | 1.02 | -1.01 | 1.01 | 1.01 | 1.06 | 1.03 | 1.0 |
| actin polymerization or depolymerization | 10 | -1.07 | 1.03 | 1.37 | 1.11 | 1.28 | -1.26 | -1.14 |
| negative regulation of DNA replication | 10 | -1.21 | 1.1 | 1.22 | 1.13 | 1.36 | -1.2 | 1.22 |
| protein phosphatase 1 binding | 25 | 1.23 | -1.0 | 1.19 | 1.06 | 1.18 | 1.25 | 1.15 |
| hedgehog receptor activity | 5 | 1.99 | 1.03 | 1.37 | -1.02 | 1.31 | 2.39 | 1.55 |
| positive regulation of transcription elongation, DNA-dependent | 11 | 1.03 | 1.26 | 1.29 | -1.01 | 1.22 | 1.24 | 1.55 |
| protein tyrosine phosphatase activator activity | 3 | 1.39 | 1.03 | 1.11 | 1.1 | 1.16 | 1.44 | 1.03 |
| response to carbamate | 1 | 2.27 | 1.23 | -5.17 | -1.59 | -2.2 | -9.9 | 9.42 |
| response to cyclodiene | 1 | 1.0 | 1.06 | 1.02 | -1.01 | -1.21 | 1.06 | -1.07 |
| response to DDT | 1 | -5.76 | 1.9 | -16.55 | 1.25 | -6.77 | -4.39 | 3.09 |
| response to organophosphorus | 2 | -1.59 | 1.53 | -9.25 | -1.13 | -3.86 | -6.59 | 5.39 |
| methyltransferase activity | 99 | 1.29 | 1.12 | 1.16 | 1.16 | 1.2 | 1.2 | 1.04 |
| C-methyltransferase activity | 2 | -1.09 | 1.1 | 1.25 | 1.35 | 1.18 | 1.22 | -1.29 |
| N-methyltransferase activity | 28 | 1.25 | 1.16 | 1.31 | 1.09 | 1.25 | 1.3 | 1.16 |
| O-methyltransferase activity | 8 | 2.13 | -1.36 | -1.3 | -1.24 | 1.01 | 1.32 | 1.14 |
| S-methyltransferase activity | 3 | -1.85 | 1.29 | -1.98 | 1.47 | 1.67 | -1.31 | -1.12 |
| RNA methyltransferase activity | 21 | 1.33 | 1.19 | 1.23 | 1.28 | 1.27 | 1.08 | 1.02 |
| mRNA methyltransferase activity | 3 | 1.33 | 1.16 | 1.39 | 1.35 | 1.34 | 1.1 | 1.01 |
| tRNA methyltransferase activity | 7 | 1.13 | 1.18 | -1.0 | 1.31 | 1.1 | -1.11 | -1.05 |
| tRNA (guanine-N7-)-methyltransferase activity | 1 | 1.71 | 1.4 | 1.24 | 1.52 | 1.28 | 1.04 | 1.1 |
| succinate dehydrogenase (ubiquinone) activity | 4 | -1.04 | 1.01 | 1.45 | -1.06 | -1.01 | 1.58 | -1.36 |
| signalosome | 8 | 1.14 | 1.05 | 1.18 | 1.23 | 1.23 | -1.06 | -1.08 |
| glycogen phosphorylase activity | 1 | 2.98 | -1.08 | 1.5 | -1.31 | 1.12 | 4.86 | 1.33 |
| RNA-dependent ATPase activity | 38 | 1.22 | 1.21 | 1.3 | 1.2 | 1.31 | 1.37 | 1.16 |
| poly-pyrimidine tract binding | 11 | 1.49 | 1.11 | 1.46 | 1.08 | 1.34 | 1.6 | 1.2 |
| neuropeptide receptor activity | 44 | 1.12 | -1.07 | 1.02 | -1.07 | -1.04 | 1.16 | 1.01 |
| eukaryotic initiation factor 4E binding | 1 | 1.26 | 1.58 | 1.86 | 1.05 | 1.71 | 1.8 | 2.46 |
| metalloendopeptidase inhibitor activity | 1 | 8.96 | -2.17 | 1.12 | -1.71 | -1.16 | 3.1 | 2.84 |
| RNA guanylyltransferase activity | 2 | 1.49 | 1.24 | 1.25 | 1.25 | 1.15 | 1.46 | 1.15 |
| tRNA guanylyltransferase activity | 1 | 1.31 | 1.2 | 1.2 | 1.21 | 1.12 | 1.43 | -1.01 |
| UDP-glycosyltransferase activity | 97 | -1.16 | -1.03 | -1.06 | 1.0 | 1.11 | -1.15 | 1.15 |
| phosphatidate phosphatase activity | 9 | 2.18 | -1.22 | -1.33 | -1.14 | 1.06 | -1.03 | 1.06 |
| vitellogenin receptor activity | 1 | 1.15 | -1.08 | 1.04 | 1.21 | 1.31 | 1.09 | -1.03 |
| ferrous iron binding | 3 | -1.78 | 1.04 | 1.19 | 1.02 | -1.06 | -2.25 | -1.21 |
| ferric iron binding | 6 | 1.86 | -1.38 | -1.24 | -1.34 | -1.27 | 1.74 | 1.1 |
| heparin binding | 10 | -1.15 | 1.06 | 1.17 | 1.03 | 1.27 | -1.23 | 1.15 |
| steroid metabolic process | 25 | 1.6 | -1.17 | 1.06 | -1.05 | -1.04 | 1.23 | 1.07 |
| cholesterol metabolic process | 4 | 1.71 | 1.17 | -1.04 | 1.01 | 1.09 | 1.45 | 1.5 |
| ecdysone metabolic process | 9 | 1.02 | 1.03 | 1.05 | 1.08 | 1.14 | 1.15 | 1.0 |
| bile acid metabolic process | 1 | -1.02 | 1.14 | 1.14 | 1.39 | 1.15 | 1.04 | -1.06 |
| androgen metabolic process | 1 | 1.03 | -1.04 | 1.2 | -1.01 | 1.0 | 2.11 | -1.07 |
| estrogen metabolic process | 1 | 1.03 | -1.04 | 1.2 | -1.01 | 1.0 | 2.11 | -1.07 |
| protein alkylation | 32 | 1.29 | 1.19 | 1.36 | 1.15 | 1.36 | 1.34 | 1.2 |
| protein dealkylation | 7 | 1.18 | 1.25 | 1.72 | 1.13 | 1.59 | 1.68 | 1.3 |
| spermine metabolic process | 2 | 1.12 | 1.21 | 1.31 | 1.22 | 1.2 | 1.92 | 1.01 |
| spermidine metabolic process | 4 | 1.11 | 1.15 | 1.16 | 1.22 | 1.17 | 1.34 | -1.08 |
| cell death | 153 | 1.07 | 1.04 | 1.16 | 1.06 | 1.25 | 1.12 | 1.14 |
| programmed cell death | 148 | 1.06 | 1.05 | 1.17 | 1.06 | 1.26 | 1.12 | 1.14 |
| cytolysis | 1 | 2.51 | -2.79 | -2.4 | 2.63 | 1.21 | 1.32 | -1.1 |
| defense response to Gram-positive bacterium | 32 | 1.3 | -1.43 | -1.38 | -1.33 | -1.3 | -1.1 | 1.27 |
| defense response to Gram-negative bacterium | 32 | 1.66 | -1.16 | -1.17 | -1.19 | -1.05 | 1.16 | 1.24 |
| tyramine receptor activity | 2 | -1.26 | 1.29 | -1.12 | 1.24 | -1.01 | -1.45 | 1.11 |
| G-protein coupled amine receptor activity | 18 | 1.01 | 1.03 | 1.15 | 1.04 | 1.0 | -1.07 | -1.05 |
| ecdysone receptor holocomplex | 4 | 1.21 | 1.23 | 1.34 | 1.21 | 1.4 | 1.24 | 1.26 |
| repressor ecdysone receptor complex | 2 | 1.38 | 1.15 | 1.29 | 1.1 | 1.25 | 1.35 | 1.22 |
| exopeptidase activity | 98 | -1.97 | -1.3 | -1.45 | 1.04 | -1.09 | -1.69 | -1.59 |
| omega peptidase activity | 7 | 1.51 | -1.0 | -1.02 | 1.11 | 1.16 | 1.09 | 1.0 |
| oligosaccharyltransferase complex | 2 | -1.97 | 1.27 | 1.32 | 1.05 | 1.26 | -1.0 | 1.71 |
| tRNA-specific adenosine deaminase activity | 1 | -1.25 | 1.12 | 1.26 | -1.01 | -1.04 | -1.21 | -1.06 |
| nucleotidase activity | 10 | -1.98 | 1.08 | -1.56 | 1.17 | -1.11 | -1.91 | -1.09 |
| 5'-nucleotidase activity | 7 | -2.99 | 1.07 | -1.74 | 1.18 | -1.15 | -2.4 | -1.14 |
| ecdysis-triggering hormone activity | 2 | -1.05 | 1.04 | -1.07 | -1.01 | -1.12 | -1.1 | -1.01 |
| head involution | 37 | 1.14 | 1.08 | 1.31 | 1.05 | 1.26 | 1.12 | 1.32 |
| 3-oxoacid CoA-transferase activity | 1 | -1.42 | 1.61 | -1.39 | -1.13 | -1.97 | 3.73 | -1.22 |
| allatostatin receptor activity | 5 | -1.14 | -1.08 | 1.25 | -1.02 | 1.16 | 1.25 | -1.31 |
| importin-alpha export receptor activity | 1 | 1.02 | 1.3 | 1.9 | 1.26 | 1.85 | 1.34 | 1.32 |
| pyrimidine-specific mismatch base pair DNA N-glycosylase activity | 1 | -1.08 | 1.06 | -1.04 | 1.0 | -1.17 | 1.06 | -1.14 |
| Mo-molybdopterin cofactor sulfurase activity | 3 | -1.23 | 1.5 | -1.02 | 1.17 | -1.21 | -2.29 | 1.26 |
| poly(U) RNA binding | 4 | 1.37 | 1.1 | 1.19 | 1.12 | 1.19 | 1.46 | 1.12 |
| JAK pathway signal transduction adaptor activity | 1 | -2.42 | 1.28 | 1.48 | 1.16 | 1.17 | -1.51 | 1.15 |
| zinc ion binding | 854 | 1.03 | 1.05 | 1.1 | 1.09 | 1.18 | 1.04 | -1.01 |
| secondary active sulfate transmembrane transporter activity | 9 | -1.49 | -1.12 | -1.96 | -1.28 | -1.37 | 2.2 | -1.04 |
| sulfate transport | 4 | -1.28 | -1.14 | 1.13 | -1.09 | 1.33 | 2.21 | -1.19 |
| calcium, potassium:sodium antiporter activity | 5 | -1.42 | 1.17 | -1.18 | 1.1 | -1.14 | -1.59 | -1.02 |
| gamma-tubulin ring complex | 5 | 1.67 | -1.04 | 1.23 | 1.09 | 1.31 | 1.41 | 1.1 |
| gamma-tubulin small complex | 4 | 1.69 | 1.04 | 1.32 | 1.06 | 1.39 | 1.77 | 1.19 |
| protein methyltransferase activity | 28 | 1.22 | 1.17 | 1.33 | 1.13 | 1.3 | 1.24 | 1.15 |
| regulation of G-protein coupled receptor protein signaling pathway | 25 | 1.29 | 1.06 | 1.24 | 1.1 | 1.16 | 1.15 | 1.13 |
| cohesin complex | 11 | 1.22 | 1.11 | 1.37 | 1.14 | 1.35 | 1.4 | 1.12 |
| cohesin core heterodimer | 1 | 2.72 | 1.09 | 2.08 | 1.56 | 2.05 | 2.2 | 1.21 |
| sulfonylurea receptor activity | 1 | 2.13 | 1.07 | 1.24 | -1.07 | 1.02 | 1.64 | 1.13 |
| ATP-sensitive potassium channel complex | 1 | 2.13 | 1.07 | 1.24 | -1.07 | 1.02 | 1.64 | 1.13 |
| cell proliferation | 156 | 1.11 | 1.12 | 1.26 | 1.11 | 1.32 | 1.15 | 1.12 |
| positive regulation of cell proliferation | 30 | 1.16 | 1.02 | 1.29 | 1.1 | 1.47 | 1.23 | 1.2 |
| negative regulation of cell proliferation | 40 | 1.43 | 1.07 | 1.41 | 1.04 | 1.41 | 1.49 | 1.16 |
| insulin receptor signaling pathway | 19 | -1.09 | 1.12 | 1.15 | 1.13 | 1.17 | 1.07 | 1.18 |
| protein serine/threonine phosphatase complex | 28 | 1.25 | 1.08 | 1.34 | 1.05 | 1.22 | 1.54 | 1.16 |
| F-actin capping protein complex | 2 | -1.38 | -1.03 | 2.18 | 1.01 | 1.59 | -1.39 | -1.04 |
| acetylcholine metabolic process | 3 | 1.5 | 1.03 | -1.96 | -1.22 | -1.24 | -2.29 | 2.08 |
| acetylcholine biosynthetic process | 2 | 1.23 | -1.06 | -1.21 | -1.07 | 1.07 | -1.1 | -1.02 |
| torso signaling pathway | 33 | 1.26 | 1.03 | 1.24 | 1.02 | 1.16 | 1.22 | 1.22 |
| calcium- and calmodulin-responsive adenylate cyclase activity | 1 | 4.63 | 2.09 | 1.35 | -1.43 | -1.28 | 1.63 | 2.43 |
| spermidine biosynthetic process | 3 | 1.26 | 1.15 | 1.19 | 1.19 | 1.22 | 1.53 | -1.02 |
| 3'-5'-exodeoxyribonuclease activity | 5 | -1.07 | 1.09 | 1.04 | -1.0 | 1.17 | 1.05 | -1.01 |
| single-stranded DNA specific exodeoxyribonuclease activity | 1 | 1.7 | -1.05 | -1.07 | -1.09 | -1.18 | 1.33 | 1.14 |
| intracellular mRNA localization | 55 | 1.09 | 1.16 | 1.32 | 1.14 | 1.25 | 1.23 | 1.17 |
| isoprenoid biosynthetic process | 12 | 1.73 | -1.18 | 1.02 | -1.16 | 1.13 | 1.29 | 1.36 |
| isoprenoid catabolic process | 4 | -1.67 | 1.21 | -1.1 | -1.05 | -1.31 | -3.02 | 1.26 |
| DNA binding, bending | 6 | 1.07 | 1.1 | 1.11 | 1.22 | 1.35 | -1.22 | 1.04 |
| female germline ring canal formation, actin assembly | 2 | 1.08 | 1.2 | 1.17 | -1.17 | 1.32 | 2.65 | 1.46 |
| caspase complex | 2 | -3.83 | 1.28 | 1.42 | 1.35 | 1.72 | -4.2 | -3.4 |
| eukaryotic translation initiation factor 4F complex | 11 | 1.11 | 1.13 | 1.2 | 1.02 | 1.09 | 1.09 | 1.35 |
| associative learning | 55 | 1.1 | 1.08 | 1.18 | 1.01 | 1.2 | 1.31 | 1.21 |
| structural constituent of muscle | 14 | 2.28 | -1.15 | 1.21 | -1.08 | 1.27 | 1.81 | -1.09 |
| voltage-gated anion channel activity | 9 | -1.16 | 1.09 | 1.05 | 1.07 | -1.0 | 1.42 | 1.04 |
| double-stranded DNA specific exodeoxyribonuclease activity | 2 | 1.46 | -1.04 | -1.04 | 1.08 | 1.25 | 1.21 | 1.13 |
| single-stranded DNA specific 3'-5' exodeoxyribonuclease activity | 1 | 1.7 | -1.05 | -1.07 | -1.09 | -1.18 | 1.33 | 1.14 |
| double-stranded DNA specific 3'-5' exodeoxyribonuclease activity | 2 | 1.46 | -1.04 | -1.04 | 1.08 | 1.25 | 1.21 | 1.13 |
| 7S RNA binding | 6 | 1.07 | 1.41 | 1.11 | 1.17 | 1.04 | -1.21 | 1.28 |
| gurken-activated receptor activity | 1 | 2.54 | -1.11 | -1.09 | 1.14 | 1.59 | 3.23 | 1.89 |
| gurken receptor signaling pathway | 4 | 2.03 | 1.05 | 1.12 | 1.25 | 1.3 | 1.58 | 1.1 |
| meiotic G2/MI transition | 5 | -1.32 | 1.33 | 1.45 | 1.2 | 1.64 | 1.0 | 1.18 |
| structural constituent of vitelline membrane | 5 | -1.04 | 1.0 | -1.05 | -1.03 | -1.16 | -1.05 | -1.08 |
| gurken receptor binding | 2 | 3.37 | -1.03 | 1.31 | 1.11 | 1.37 | 1.92 | 1.47 |
| protein prenyltransferase activity | 7 | -1.61 | 1.12 | 1.15 | 1.12 | 1.17 | -1.52 | 1.36 |
| protein transmembrane transporter activity | 33 | 1.03 | 1.14 | 1.21 | 1.1 | 1.12 | 1.16 | -1.01 |
| Ral guanyl-nucleotide exchange factor activity | 1 | 8.19 | -2.13 | 1.05 | -1.9 | 1.69 | 3.12 | 2.66 |
| larval feeding behavior | 14 | -1.24 | 1.04 | 1.52 | -1.07 | -1.05 | -1.09 | 1.18 |
| cation transmembrane transporter activity | 354 | -1.17 | -1.05 | -1.15 | -1.12 | -1.07 | 1.16 | -1.11 |
| methyl-CpG binding | 1 | 2.49 | 1.08 | 1.5 | 1.28 | 1.63 | 1.64 | 1.5 |
| ionotropic glutamate receptor complex | 5 | -1.46 | 1.04 | 1.09 | -1.04 | -1.01 | -1.5 | 1.07 |
| protein tyrosine/threonine phosphatase activity | 1 | 1.33 | 1.23 | 1.28 | 1.29 | 1.18 | 1.21 | 1.19 |
| low voltage-gated calcium channel activity | 1 | -1.24 | 1.06 | -1.0 | 1.0 | -1.1 | 1.0 | -1.05 |
| endosome to lysosome transport | 7 | 1.05 | 1.14 | 1.56 | 1.12 | 1.35 | 1.31 | 1.19 |
| histone mRNA metabolic process | 9 | 1.42 | 1.07 | 1.16 | 1.23 | 1.3 | 1.29 | 1.06 |
| female germline ring canal stabilization | 4 | -1.41 | 1.17 | 1.28 | 1.2 | 1.33 | -1.35 | 1.02 |
| gamma-butyrobetaine dioxygenase activity | 4 | -2.34 | -1.03 | -1.77 | -1.06 | -1.07 | -1.32 | 1.02 |
| determination of adult lifespan | 113 | 1.24 | 1.12 | 1.11 | -1.02 | 1.16 | 1.36 | 1.25 |
| behavioral response to cocaine | 9 | 1.31 | 1.15 | 1.55 | 1.04 | 1.35 | 1.53 | 1.26 |
| adult feeding behavior | 7 | 1.32 | -1.86 | -1.97 | -2.12 | -2.07 | -1.51 | 2.24 |
| adult locomotory behavior | 50 | 1.11 | 1.02 | 1.21 | 1.0 | 1.17 | 1.25 | 1.14 |
| larval locomotory behavior | 18 | 1.01 | 1.12 | 1.09 | -1.24 | -1.15 | -1.09 | 1.14 |
| glial cell migration | 26 | 1.24 | 1.07 | 1.41 | 1.1 | 1.38 | 1.25 | 1.08 |
| negative regulation of antimicrobial humoral response | 4 | 1.35 | 1.1 | 1.46 | 1.15 | 1.8 | 1.12 | 1.2 |
| MAP kinase kinase kinase kinase activity | 1 | -1.25 | 1.21 | 1.4 | 1.19 | 1.07 | 1.11 | 1.44 |
| microtubule severing | 6 | -1.04 | 1.14 | 1.1 | 1.14 | 1.18 | 1.01 | 1.05 |
| katanin complex | 4 | 1.09 | 1.1 | 1.08 | 1.22 | 1.19 | -1.04 | 1.02 |
| RNA polymerase II carboxy-terminal domain kinase activity | 5 | 1.49 | 1.22 | 1.54 | 1.24 | 1.64 | 1.52 | 1.29 |
| germ cell migration | 42 | 1.28 | 1.05 | 1.21 | -1.02 | 1.39 | 1.39 | 1.26 |
| olfactory learning | 49 | 1.08 | 1.08 | 1.17 | -1.0 | 1.22 | 1.27 | 1.23 |
| asymmetric cell division | 66 | 1.06 | 1.13 | 1.27 | 1.08 | 1.35 | 1.05 | 1.13 |
| maternal determination of anterior/posterior axis, embryo | 66 | 1.14 | 1.17 | 1.4 | 1.11 | 1.34 | 1.36 | 1.21 |
| regulation of bicoid mRNA localization | 2 | -1.31 | 1.2 | 1.89 | -1.13 | 1.39 | 2.71 | 1.31 |
| regulation of cell shape | 104 | 1.18 | 1.11 | 1.41 | 1.04 | 1.38 | 1.42 | 1.29 |
| regulation of cell size | 48 | 1.12 | 1.2 | 1.33 | 1.06 | 1.31 | 1.38 | 1.25 |
| chitin-based embryonic cuticle biosynthetic process | 18 | 1.73 | -1.03 | 1.1 | -1.15 | 1.04 | 1.97 | 1.4 |
| larval chitin-based cuticle development | 12 | 1.05 | -1.05 | 1.07 | -1.09 | 1.02 | -1.12 | 1.35 |
| pupal chitin-based cuticle development | 1 | 14.19 | -4.21 | -4.1 | -3.94 | -3.79 | 21.36 | -4.46 |
| adult chitin-based cuticle development | 10 | 1.01 | -1.17 | -1.08 | -1.07 | -1.12 | -1.07 | 1.3 |
| axon ensheathment | 6 | 1.77 | -1.07 | 1.38 | -1.13 | 1.24 | 3.0 | 1.46 |
| sialyltransferase activity | 1 | 1.19 | 1.12 | 1.07 | 1.08 | 1.03 | 1.22 | -1.05 |
| O-acyltransferase activity | 20 | -1.13 | -1.08 | -1.14 | 1.21 | 1.65 | -1.68 | -1.37 |
| acetylglucosaminyltransferase activity | 26 | 1.0 | 1.15 | 1.12 | 1.08 | 1.33 | 1.22 | 1.29 |
| acetylgalactosaminyltransferase activity | 18 | -1.14 | -1.09 | -1.18 | -1.18 | -1.19 | -1.47 | 1.62 |
| light-induced release of internally sequestered calcium ion | 1 | -1.32 | -1.06 | -1.2 | 1.5 | -1.29 | -1.26 | -1.29 |
| galactosyltransferase activity | 23 | -1.33 | -1.04 | -1.03 | 1.03 | 1.03 | -1.42 | 1.2 |
| thioredoxin peroxidase activity | 6 | -1.25 | 1.02 | -1.13 | 1.12 | 1.03 | -1.35 | -1.22 |
| mechanically-gated ion channel activity | 1 | -1.05 | 1.0 | -1.06 | -1.03 | -1.14 | 1.01 | 1.95 |
| IkappaB kinase activity | 3 | 1.21 | -1.06 | 1.39 | 1.12 | 1.46 | -1.08 | -1.04 |
| IkappaB kinase complex | 3 | 1.21 | -1.06 | 1.39 | 1.12 | 1.46 | -1.08 | -1.04 |
| steroid hydroxylase activity | 5 | -1.11 | -1.12 | 1.02 | -1.03 | -1.09 | -1.02 | -1.19 |
| gonad development | 42 | 1.01 | 1.04 | 1.05 | 1.07 | 1.24 | -1.01 | 1.0 |
| chaeta morphogenesis | 42 | 1.01 | 1.11 | 1.31 | 1.05 | 1.22 | 1.07 | 1.18 |
| 3'-5' exonuclease activity | 25 | 1.2 | 1.14 | 1.2 | 1.19 | 1.29 | 1.17 | 1.03 |
| 5'-3' exonuclease activity | 2 | -1.11 | 1.53 | 1.16 | 1.33 | 1.34 | -1.11 | 1.19 |
| CoA-transferase activity | 2 | -1.15 | 1.38 | -1.15 | 1.04 | -1.3 | 1.87 | -1.04 |
| 4-hydroxybutyrate CoA-transferase activity | 1 | 1.07 | 1.18 | 1.06 | 1.23 | 1.16 | -1.07 | 1.13 |
| 4-hydroxybenzoate octaprenyltransferase activity | 1 | 1.04 | 1.26 | 1.1 | 1.28 | -1.02 | -1.97 | -1.12 |
| CDP-alcohol phosphotransferase activity | 1 | -10.11 | 1.17 | -1.48 | 1.2 | 1.57 | -5.09 | -1.84 |
| delta5-delta2,4-dienoyl-CoA isomerase activity | 2 | 1.86 | -1.05 | 1.02 | 1.19 | 1.39 | 1.16 | -1.03 |
| fucosyltransferase activity | 7 | -1.17 | 1.08 | -1.01 | 1.15 | 1.23 | -1.03 | 1.02 |
| protein-N-terminal asparagine amidohydrolase activity | 1 | 6.19 | -1.42 | -1.44 | -1.3 | -1.36 | 1.94 | 1.84 |
| RNA lariat debranching enzyme activity | 2 | 1.26 | 1.13 | 1.23 | 1.25 | 1.39 | 1.4 | 1.02 |
| CTD phosphatase activity | 2 | -1.16 | 1.24 | 1.23 | 1.2 | 1.16 | 1.07 | 1.3 |
| beta-glucosidase activity | 1 | -1.02 | 1.14 | 1.14 | 1.39 | 1.15 | 1.04 | -1.06 |
| glycoprotein 6-alpha-L-fucosyltransferase activity | 1 | 1.63 | 1.17 | -1.53 | -1.05 | -1.49 | 4.17 | 1.53 |
| 2-polyprenyl-6-methoxy-1,4-benzoquinone methyltransferase activity | 2 | -1.09 | 1.1 | 1.25 | 1.35 | 1.18 | 1.22 | -1.29 |
| protein kinase C inhibitor activity | 2 | 1.45 | 1.07 | 1.48 | 1.05 | 1.45 | 1.77 | 1.84 |
| betaine transport | 2 | 1.34 | 1.32 | -1.91 | 1.22 | 1.14 | -1.7 | -1.23 |
| ribonuclease inhibitor activity | 2 | 1.78 | 1.2 | 1.19 | -1.04 | 1.08 | 2.12 | 1.24 |
| phosphatidylethanolamine binding | 8 | 1.31 | -1.34 | -1.15 | -1.46 | -1.4 | 1.2 | 1.09 |
| selenium binding | 4 | 1.26 | 1.16 | 1.54 | 1.18 | 1.1 | 1.17 | -1.12 |
| vitamin E binding | 10 | -2.36 | -2.69 | -2.86 | -1.36 | -1.28 | -1.63 | -1.06 |
| JUN kinase binding | 1 | -1.08 | 1.01 | 1.18 | 1.12 | 1.07 | -1.04 | 1.06 |
| heterogeneous nuclear ribonucleoprotein complex | 7 | 1.06 | 1.34 | 1.44 | 1.06 | 1.21 | 1.65 | 1.29 |
| inositol-1,4,5-trisphosphate 3-kinase activity | 5 | 1.28 | 1.12 | 1.04 | -1.27 | 1.01 | 1.27 | 1.33 |
| 3'(2'),5'-bisphosphate nucleotidase activity | 2 | 1.61 | 1.02 | -1.31 | 1.13 | -1.08 | 1.03 | 1.2 |
| 3-hydroxyisobutyrate dehydrogenase activity | 2 | -1.14 | 1.35 | 1.05 | 1.01 | 1.01 | 1.03 | 1.42 |
| phosphofructokinase activity | 2 | 1.69 | -1.28 | 1.45 | -1.01 | 1.01 | 4.15 | 1.1 |
| CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase activity | 1 | 1.09 | 1.12 | -1.13 | 1.16 | 1.11 | -1.64 | -1.3 |
| D-aspartate oxidase activity | 2 | -2.82 | -1.69 | -2.22 | 1.7 | 1.95 | -1.63 | -1.83 |
| GDP-mannose 4,6-dehydratase activity | 1 | -3.57 | 1.21 | 1.13 | 1.39 | 1.5 | -2.8 | -1.34 |
| L-ascorbate oxidase activity | 1 | -1.08 | -1.06 | -1.06 | -1.17 | -1.14 | -1.13 | -1.1 |
| N-acetylglucosamine-6-phosphate deacetylase activity | 1 | 1.05 | -1.01 | -1.53 | 1.51 | 1.2 | -1.51 | -1.4 |
| N-acetylglucosamine-6-sulfatase activity | 2 | 4.3 | -1.11 | 1.07 | -1.47 | 1.62 | 8.46 | 1.92 |
| alanine-glyoxylate transaminase activity | 3 | -2.83 | 1.14 | -4.47 | -1.13 | -1.51 | -1.62 | 1.13 |
| alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity | 2 | 1.98 | -1.1 | 1.19 | -1.09 | 1.23 | 2.85 | 1.26 |
| alpha-1,6-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity | 1 | 3.56 | -1.43 | -1.34 | -1.25 | -1.02 | 7.35 | 1.28 |
| alpha-N-acetylgalactosaminidase activity | 1 | 1.06 | -1.01 | -1.01 | 1.04 | -1.07 | -1.04 | 1.13 |
| carnitine O-octanoyltransferase activity | 1 | 2.33 | -1.31 | -2.71 | 1.32 | 2.24 | -2.61 | 1.02 |
| chondroitin 6-sulfotransferase activity | 1 | 1.06 | 1.47 | 1.34 | 1.13 | 1.02 | -1.02 | 1.04 |
| dTDP-glucose 4,6-dehydratase activity | 1 | 2.45 | 1.09 | 2.09 | -1.01 | 1.2 | 1.56 | 1.31 |
| glycogenin glucosyltransferase activity | 2 | 7.0 | -1.59 | 1.16 | -1.42 | 1.29 | 4.38 | -1.04 |
| [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity | 2 | 1.57 | 1.14 | -1.02 | 1.0 | 1.08 | -1.33 | -1.08 |
| histone-arginine N-methyltransferase activity | 2 | 1.08 | 1.15 | 1.55 | 1.17 | 1.64 | 1.08 | 1.14 |
| isovaleryl-CoA dehydrogenase activity | 1 | 1.14 | 1.39 | 1.15 | 1.11 | 1.04 | 1.05 | 1.01 |
| hydroquinone:oxygen oxidoreductase activity | 2 | -3.8 | -3.2 | -3.92 | -3.61 | -4.08 | -3.9 | 3.04 |
| palmitoyl-(protein) hydrolase activity | 2 | 2.75 | 1.12 | 1.29 | 1.19 | 1.61 | 1.16 | 1.22 |
| procollagen-lysine 5-dioxygenase activity | 2 | 1.13 | 1.08 | 1.05 | -1.04 | 1.19 | 1.95 | 1.17 |
| protein-tyrosine sulfotransferase activity | 1 | -1.29 | 1.27 | 1.45 | -1.08 | 1.08 | 1.07 | 2.33 |
| queuine tRNA-ribosyltransferase activity | 2 | -1.08 | 1.07 | 1.01 | 1.12 | 1.08 | -1.67 | -1.22 |
| sarcosine dehydrogenase activity | 1 | 1.0 | 1.04 | -1.1 | -1.02 | -1.03 | 1.1 | 1.13 |
| sulfite oxidase activity | 1 | -1.72 | 1.15 | -1.04 | 1.56 | -1.05 | 1.22 | -1.45 |
| transaminase activity | 14 | -1.03 | 1.19 | -1.47 | -1.11 | -1.05 | 1.04 | 1.34 |
| sulfuric ester hydrolase activity | 8 | 1.82 | -1.14 | -1.15 | -1.09 | 1.33 | 2.75 | 1.13 |
| diphosphoinositol-polyphosphate diphosphatase activity | 1 | 1.43 | 1.14 | 1.21 | 1.02 | 1.13 | 1.54 | 1.16 |
| gamma-glutamyl carboxylase activity | 1 | 1.13 | 1.29 | 1.08 | 1.08 | -1.09 | -1.02 | 1.13 |
| arsenite secondary active transmembrane transporter activity | 1 | 1.35 | 1.29 | 1.36 | 1.08 | 1.19 | 1.34 | 1.11 |
| cAMP metabolic process | 7 | 1.35 | 1.03 | 1.31 | -1.01 | 1.11 | 1.49 | 1.22 |
| translation activator activity | 1 | 1.37 | 1.21 | 2.02 | -1.05 | 1.64 | 2.25 | 1.61 |
| protoheme IX farnesyltransferase activity | 1 | -1.18 | 1.24 | 1.2 | 1.18 | -1.04 | 1.79 | -1.68 |
| mannan endo-1,6-alpha-mannosidase activity | 1 | 1.08 | 1.1 | 1.1 | 1.25 | 1.18 | 1.04 | 1.09 |
| phospholipid scrambling | 2 | 2.57 | -1.14 | 1.03 | -1.07 | 2.13 | 1.16 | 1.16 |
| UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity | 7 | 1.21 | 1.06 | 1.08 | 1.06 | -1.01 | 1.55 | 1.16 |
| glycine binding | 1 | 2.28 | 1.03 | 1.38 | -1.44 | -1.34 | 1.53 | 1.42 |
| glutamate binding | 4 | 1.16 | 1.11 | 1.12 | -1.06 | -1.11 | 1.09 | 1.14 |
| thienylcyclohexylpiperidine binding | 1 | 2.28 | 1.03 | 1.38 | -1.44 | -1.34 | 1.53 | 1.42 |
| melatonin receptor activity | 1 | 1.09 | 1.23 | 3.2 | 1.05 | 2.53 | 1.91 | 1.95 |
| benzodiazepine receptor activity | 1 | -1.12 | -1.26 | 1.76 | 1.06 | 1.23 | -1.31 | -1.61 |
| monoamine transmembrane transporter activity | 3 | 2.07 | -1.32 | -1.3 | -1.21 | -1.34 | 1.32 | -1.28 |
| sucrose:hydrogen symporter activity | 1 | 2.52 | -1.26 | -1.18 | -1.09 | -1.25 | 2.25 | 1.03 |
| sodium:iodide symporter activity | 7 | 1.13 | -1.12 | -1.82 | -1.93 | -1.54 | -1.43 | 1.18 |
| bile acid:sodium symporter activity | 2 | -3.13 | 1.41 | -2.05 | 1.29 | -3.5 | -4.18 | -1.28 |
| anion transmembrane transporter activity | 90 | -1.43 | -1.05 | -1.42 | -1.25 | -1.3 | 1.08 | -1.17 |
| sodium:bicarbonate symporter activity | 1 | 1.2 | -1.03 | 1.07 | -1.04 | 1.13 | 1.1 | 1.23 |
| sodium:potassium:chloride symporter activity | 3 | -1.68 | -1.44 | 1.14 | -1.49 | -1.04 | 2.8 | -1.1 |
| secondary active organic cation transmembrane transporter activity | 24 | -1.27 | -1.38 | -1.63 | -1.24 | -1.0 | 2.31 | -1.46 |
| organic anion transmembrane transporter activity | 8 | -2.06 | 1.07 | -1.1 | -1.04 | 1.14 | -1.35 | 1.33 |
| sucrose transmembrane transporter activity | 1 | 2.52 | -1.26 | -1.18 | -1.09 | -1.25 | 2.25 | 1.03 |
| folic acid transporter activity | 1 | -1.3 | -1.21 | 1.08 | -1.14 | 1.42 | 1.82 | -1.08 |
| reduced folate carrier activity | 3 | -2.47 | 1.26 | -3.59 | 2.04 | -1.22 | -1.96 | -2.06 |
| ammonium transmembrane transporter activity | 1 | -1.13 | -1.02 | -1.13 | -1.06 | -1.15 | -1.01 | -1.16 |
| L-ascorbate:sodium symporter activity | 1 | 1.92 | -5.29 | -3.19 | -3.44 | 7.65 | 2.58 | 1.23 |
| acetyl-CoA transporter activity | 1 | -1.28 | 1.24 | -1.24 | 1.18 | -1.13 | -1.69 | 1.59 |
| sodium-dependent multivitamin transmembrane transporter activity | 5 | 1.01 | 1.09 | -2.28 | -1.32 | 1.1 | -1.3 | 1.01 |
| glucose 6-phosphate:phosphate antiporter activity | 1 | 1.19 | 1.35 | 1.39 | 1.1 | 1.22 | 1.13 | 2.76 |
| phosphatidylcholine transporter activity | 3 | 3.03 | -1.11 | 1.31 | -1.04 | 1.16 | 1.62 | 1.22 |
| phosphatidylinositol transporter activity | 3 | 2.87 | -1.14 | 1.55 | -1.02 | 1.46 | 1.97 | 1.15 |
| taste receptor activity | 55 | -1.07 | 1.01 | -1.03 | 1.01 | -1.01 | -1.07 | -1.07 |
| G-protein coupled peptide receptor activity | 44 | 1.12 | -1.07 | 1.02 | -1.07 | -1.04 | 1.16 | 1.01 |
| riboflavin kinase activity | 1 | 1.06 | 1.33 | 1.06 | -1.02 | -1.41 | 1.22 | 1.19 |
| N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity | 3 | -3.66 | 1.03 | -1.66 | 1.15 | 1.31 | -2.05 | -1.18 |
| oxidized purine base lesion DNA N-glycosylase activity | 2 | 1.64 | -1.01 | 1.15 | 1.12 | 1.14 | 1.33 | -1.07 |
| respiratory chain complex IV assembly | 8 | 1.05 | 1.23 | 1.36 | 1.17 | 1.1 | 1.44 | -1.28 |
| proteasome activator complex | 1 | 1.38 | 1.06 | -1.05 | 1.28 | -1.05 | 1.16 | -1.12 |
| endopeptidase activator activity | 3 | 1.37 | 1.13 | 1.14 | 1.18 | 1.05 | 1.05 | 1.17 |
| regulation of proteasomal protein catabolic process | 5 | 1.57 | 1.06 | 1.14 | 1.24 | 1.16 | 1.17 | 1.01 |
| proteasome regulatory particle, base subcomplex | 11 | 1.29 | 1.04 | 1.28 | 1.23 | 1.2 | 1.03 | 1.05 |
| proteasome regulatory particle, lid subcomplex | 8 | 1.42 | 1.1 | 1.26 | 1.18 | 1.26 | 1.0 | 1.06 |
| visual learning | 3 | 1.5 | 1.07 | 1.21 | 1.15 | 1.26 | 1.3 | 1.05 |
| fibroblast growth factor receptor signaling pathway | 10 | 1.63 | -1.12 | 2.13 | -1.03 | 1.14 | 1.56 | 1.09 |
| epidermis development | 59 | 1.32 | 1.05 | 1.35 | 1.0 | 1.31 | 1.31 | 1.3 |
| JUN kinase kinase activity | 2 | -1.25 | 1.26 | 1.38 | 1.09 | 1.35 | 1.25 | 1.38 |
| cadmium ion transmembrane transporter activity | 1 | -1.37 | 1.35 | 1.68 | 1.28 | 1.12 | -1.35 | -1.15 |
| ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism | 61 | -1.2 | 1.0 | 1.27 | -1.12 | -1.01 | 1.39 | -1.09 |
| hydrogen-exporting ATPase activity, phosphorylative mechanism | 41 | -1.23 | -1.01 | 1.31 | -1.14 | -1.07 | 1.48 | -1.19 |
| potassium-transporting ATPase activity | 8 | 1.13 | -1.11 | 1.22 | -1.35 | -1.0 | 1.53 | 1.06 |
| xenobiotic-transporting ATPase activity | 8 | -1.32 | 1.18 | 1.1 | 1.25 | 1.08 | -1.09 | 1.15 |
| protein transporter activity | 55 | 1.03 | 1.17 | 1.31 | 1.13 | 1.19 | 1.2 | 1.09 |
| microtubule-severing ATPase activity | 3 | -1.13 | 1.2 | 1.15 | 1.08 | 1.3 | 1.07 | 1.1 |
| minus-end-directed microtubule motor activity | 3 | 1.03 | 1.05 | 1.24 | 1.18 | 1.22 | 1.09 | -1.02 |
| actin filament-based movement | 2 | -1.84 | 1.42 | 2.86 | 1.0 | 1.74 | 1.92 | 1.21 |
| plus-end-directed microtubule motor activity | 6 | -1.13 | 1.06 | 1.13 | 1.16 | 1.26 | -1.02 | -1.11 |
| ATPase activity | 318 | -1.02 | 1.05 | 1.15 | 1.04 | 1.13 | 1.15 | 1.03 |
| JUN kinase phosphatase activity | 1 | -1.35 | 1.21 | 1.45 | -1.04 | 1.43 | -1.17 | 1.37 |
| regulation of synaptic growth at neuromuscular junction | 39 | -1.02 | 1.12 | 1.32 | 1.09 | 1.24 | 1.09 | 1.21 |
| mystery cell differentiation | 1 | -1.03 | 1.35 | 2.46 | 1.15 | 2.45 | 2.67 | 1.87 |
| male gonad development | 7 | -1.2 | 1.03 | -1.03 | 1.03 | 1.04 | -1.14 | -1.07 |
| female gonad development | 11 | 1.16 | 1.1 | 1.04 | -1.01 | 1.28 | 1.15 | 1.13 |
| imaginal disc-derived wing vein morphogenesis | 36 | 1.27 | 1.09 | 1.25 | 1.03 | 1.32 | 1.38 | 1.31 |
| imaginal disc-derived wing margin morphogenesis | 43 | 1.4 | 1.1 | 1.3 | 1.02 | 1.29 | 1.46 | 1.36 |
| release of cytoplasmic sequestered NF-kappaB | 1 | 1.41 | 1.35 | 1.47 | 1.02 | 1.54 | 2.41 | 1.84 |
| regulation of smoothened signaling pathway | 27 | 1.49 | 1.16 | 1.31 | 1.06 | 1.3 | 1.61 | 1.15 |
| regulation of Toll signaling pathway | 15 | 2.33 | -1.32 | -1.29 | -1.35 | -1.14 | 1.81 | 1.29 |
| regulation of Notch signaling pathway | 50 | -1.09 | 1.19 | 1.32 | 1.11 | 1.36 | 1.24 | 1.21 |
| photoreceptor cell morphogenesis | 13 | 1.11 | 1.16 | 1.43 | 1.14 | 1.34 | 1.12 | 1.26 |
| anterior/posterior axis specification, embryo | 114 | 1.21 | 1.09 | 1.29 | 1.04 | 1.25 | 1.28 | 1.24 |
| calcium-dependent protein serine/threonine phosphatase regulator activity | 4 | 1.2 | 1.0 | 1.18 | 1.05 | 1.13 | -1.01 | 1.02 |
| protein phosphatase type 1 regulator activity | 6 | 1.88 | -1.31 | 1.12 | 1.03 | 1.16 | 1.14 | 1.17 |
| protein phosphatase type 2A regulator activity | 7 | 1.44 | -1.06 | 1.23 | -1.02 | 1.16 | 2.34 | 1.17 |
| cAMP-dependent protein kinase regulator activity | 6 | -1.1 | 1.09 | 1.07 | 1.13 | 1.06 | -1.11 | -1.2 |
| protein kinase regulator activity | 43 | 1.19 | 1.13 | 1.19 | 1.14 | 1.26 | 1.06 | 1.09 |
| phosphorylase kinase regulator activity | 5 | 1.29 | 1.3 | 1.33 | 1.21 | 1.15 | 1.36 | 1.33 |
| attachment of spindle microtubules to kinetochore | 4 | -1.03 | 1.09 | 1.07 | 1.08 | 1.18 | 1.05 | 1.05 |
| alkylglycerone-phosphate synthase activity | 1 | 5.42 | -21.99 | -24.45 | -20.82 | -3.99 | -1.47 | 6.54 |
| lipid biosynthetic process | 111 | 1.08 | -1.13 | -1.05 | -1.09 | 1.07 | 1.05 | 1.09 |
| ether lipid biosynthetic process | 1 | 5.42 | -21.99 | -24.45 | -20.82 | -3.99 | -1.47 | 6.54 |
| diuretic hormone activity | 4 | -1.94 | -1.06 | 1.2 | 1.12 | 1.34 | -2.14 | -1.37 |
| pyridoxine metabolic process | 4 | 2.09 | 1.04 | -1.32 | 1.26 | 1.04 | 1.32 | 1.12 |
| pyridoxine biosynthetic process | 4 | 2.09 | 1.04 | -1.32 | 1.26 | 1.04 | 1.32 | 1.12 |
| queuosine biosynthetic process | 2 | -1.08 | 1.07 | 1.01 | 1.12 | 1.08 | -1.67 | -1.22 |
| guanosine metabolic process | 2 | -1.08 | 1.07 | 1.01 | 1.12 | 1.08 | -1.67 | -1.22 |
| 7-methylguanosine metabolic process | 2 | -1.08 | 1.07 | 1.01 | 1.12 | 1.08 | -1.67 | -1.22 |
| epsilon DNA polymerase complex | 3 | 1.73 | 1.2 | 1.21 | 1.15 | 1.32 | 1.1 | -1.16 |
| chromatin accessibility complex | 4 | 1.2 | 1.15 | 1.7 | 1.23 | 1.78 | 1.39 | 1.32 |
| induction of apoptosis by extracellular signals | 1 | 1.28 | 1.14 | -1.32 | 1.35 | 1.43 | 1.41 | 1.45 |
| induction of apoptosis by ionic changes | 2 | 1.02 | 1.0 | -1.02 | -1.06 | -1.0 | -1.03 | 1.59 |
| induction of apoptosis by hormones | 1 | 1.28 | 1.14 | -1.32 | 1.35 | 1.43 | 1.41 | 1.45 |
| induction of apoptosis by intracellular signals | 10 | 1.45 | 1.15 | 1.09 | 1.11 | 1.25 | 1.28 | 1.38 |
| DNA damage response, signal transduction resulting in induction of apoptosis | 9 | 1.53 | 1.18 | 1.1 | 1.13 | 1.27 | 1.31 | 1.44 |
| activation of pro-apoptotic gene products | 1 | 1.78 | 1.11 | 1.56 | 1.24 | 1.76 | -1.03 | 1.11 |
| urea transport | 1 | 1.41 | 1.17 | -1.02 | 1.18 | -1.05 | -1.09 | 1.18 |
| small protein activating enzyme activity | 6 | 1.19 | 1.08 | 1.24 | 1.27 | 1.39 | 1.11 | -1.07 |
| carbohydrate transport | 13 | 1.22 | -1.13 | -1.12 | -1.07 | 1.01 | 1.59 | 1.23 |
| hexose transport | 2 | -1.9 | 1.16 | -1.07 | 1.04 | -1.18 | -1.96 | 1.78 |
| rRNA methyltransferase activity | 7 | 1.35 | 1.22 | 1.24 | 1.28 | 1.26 | 1.11 | 1.02 |
| rRNA (uridine-2'-O-)-methyltransferase activity | 3 | 1.55 | 1.21 | 1.1 | 1.32 | 1.19 | 1.02 | -1.14 |
| actin filament polymerization | 6 | -1.34 | 1.04 | 1.42 | 1.14 | 1.28 | -1.56 | -1.16 |
| identical protein binding | 70 | 1.32 | 1.03 | 1.02 | -1.03 | 1.12 | 1.22 | 1.14 |
| cellular amino acid biosynthetic process | 39 | 1.02 | 1.06 | -1.29 | -1.04 | -1.12 | 1.11 | 1.06 |
| lipopolysaccharide metabolic process | 2 | 1.01 | -1.07 | -1.02 | 1.06 | 1.11 | -1.07 | -1.11 |
| phospholipid biosynthetic process | 44 | -1.11 | 1.0 | 1.16 | 1.01 | 1.28 | -1.04 | -1.11 |
| 2'-phosphotransferase activity | 1 | 3.82 | 1.09 | 1.67 | 1.1 | 1.4 | 2.3 | 1.73 |
| N-acetylmuramoyl-L-alanine amidase activity | 11 | -1.39 | -1.63 | -1.15 | 1.01 | -1.87 | -2.01 | -1.36 |
| oxidoreductase activity, acting on NADH or NADPH | 44 | 1.12 | 1.05 | 1.23 | 1.06 | 1.09 | 1.58 | -1.22 |
| S-adenosylmethionine-dependent methyltransferase activity | 52 | 1.26 | 1.1 | 1.1 | 1.12 | 1.21 | 1.16 | 1.09 |
| UDP-N-acetylmuramate dehydrogenase activity | 3 | 1.89 | -2.35 | -2.83 | -2.17 | -1.54 | -1.15 | 1.9 |
| acetaldehyde dehydrogenase (acetylating) activity | 2 | 2.78 | -1.16 | -2.38 | -2.31 | -1.44 | 1.4 | 1.22 |
| N-acylneuraminate cytidylyltransferase activity | 1 | 1.0 | -1.04 | -1.09 | -1.07 | -1.02 | 1.03 | -1.14 |
| bis(5'-nucleosyl)-tetraphosphatase activity | 1 | -1.85 | 1.12 | 1.37 | 1.12 | 1.23 | -1.02 | 1.02 |
| carbon-monoxide oxygenase activity | 1 | 2.72 | -3.32 | -2.57 | -3.03 | -1.93 | 22.15 | -2.83 |
| cardiolipin synthase activity | 1 | 1.31 | 1.29 | 1.34 | 1.27 | 1.36 | 1.03 | -1.44 |
| choline dehydrogenase activity | 15 | 1.05 | -1.22 | -1.39 | 1.07 | 1.15 | 1.16 | -1.32 |
| crossover junction endodeoxyribonuclease activity | 1 | 1.08 | 1.1 | -1.01 | 1.17 | 1.1 | -1.08 | 1.01 |
| diaminohydroxyphosphoribosylaminopyrimidine deaminase activity | 1 | 2.99 | 3.14 | -4.72 | -4.54 | -4.51 | 3.75 | 1.74 |
| DNA N-glycosylase activity | 6 | 2.07 | -1.07 | 1.03 | -1.04 | 1.05 | 1.76 | 1.02 |
| exodeoxyribonuclease I activity | 1 | 1.21 | 1.03 | 1.2 | 1.27 | 1.3 | -1.05 | 1.1 |
| exodeoxyribonuclease III activity | 1 | -1.62 | 1.33 | -1.09 | 1.09 | -1.15 | -1.69 | -1.15 |
| exoribonuclease II activity | 1 | 1.38 | 1.19 | 2.04 | 1.07 | 1.63 | 2.14 | 1.72 |
| glycerate kinase activity | 1 | 1.02 | 1.55 | -1.07 | 1.39 | -1.43 | -1.25 | -1.1 |
| glycerophosphodiester phosphodiesterase activity | 6 | -2.04 | 1.29 | -1.08 | 1.03 | -1.68 | -2.4 | 1.09 |
| glycine C-acetyltransferase activity | 1 | 1.04 | -1.18 | -1.1 | -1.3 | -1.17 | 1.44 | -1.36 |
| guanosine-3',5'-bis(diphosphate) 3'-diphosphatase activity | 1 | 1.81 | -1.03 | 1.57 | 1.17 | 1.71 | 1.48 | -1.14 |
| holo-[acyl-carrier-protein] synthase activity | 1 | 1.84 | 1.09 | 2.09 | 1.3 | 2.04 | 2.34 | 1.11 |
| homocysteine S-methyltransferase activity | 2 | -2.66 | 1.46 | -3.05 | 1.35 | 1.97 | -1.55 | -1.14 |
| mannose-phosphate guanylyltransferase activity | 2 | -2.09 | 1.14 | 1.2 | 1.25 | 1.24 | -1.89 | 1.17 |
| inositol monophosphate 1-phosphatase activity | 6 | 1.86 | -1.14 | -1.69 | -1.19 | 1.02 | 1.47 | 1.43 |
| ferredoxin-NAD(P) reductase activity | 1 | 1.15 | 1.02 | 1.12 | 1.04 | 1.0 | -1.01 | 1.01 |
| oligonucleotidase activity | 1 | 1.49 | 1.12 | 1.32 | 1.17 | 1.41 | 1.35 | 1.33 |
| oxalyl-CoA decarboxylase activity | 1 | -4.09 | -1.02 | -5.68 | 1.78 | 1.15 | -8.6 | -2.19 |
| phosphoglycolate phosphatase activity | 4 | 1.72 | 1.3 | -1.05 | 1.15 | 1.06 | 1.1 | -1.14 |
| phosphatidylcholine 1-acylhydrolase activity | 9 | -31.12 | -1.57 | -1.58 | -2.13 | -3.67 | -22.6 | -1.69 |
| site-specific DNA-methyltransferase (adenine-specific) activity | 1 | 1.54 | 1.35 | -1.17 | 1.42 | -1.06 | -1.14 | -1.22 |
| DNA-methyltransferase activity | 3 | 1.31 | 1.16 | -1.05 | 1.36 | 1.06 | -1.13 | -1.08 |
| tRNA thio-modification | 3 | 1.78 | 1.18 | 1.12 | 1.35 | 1.22 | 1.11 | -1.02 |
| sulfurtransferase activity | 4 | 1.34 | 1.04 | 1.57 | 1.22 | 1.05 | 1.11 | -1.14 |
| uridylate kinase activity | 1 | -1.35 | 1.03 | 1.12 | 1.16 | 1.1 | -1.2 | -1.23 |
| dosage compensation by hyperactivation of X chromosome | 6 | 1.52 | 1.11 | 1.3 | 1.04 | 1.23 | 1.53 | 1.43 |
| dosage compensation by inactivation of X chromosome | 1 | 1.21 | 1.08 | 1.29 | -1.08 | 1.41 | 1.2 | 1.17 |
| pentose-phosphate shunt, non-oxidative branch | 2 | 2.98 | 1.26 | -2.38 | 1.09 | -1.14 | 1.07 | 1.35 |
| catabolic process | 443 | 1.07 | -1.01 | -1.01 | 1.0 | 1.07 | 1.05 | 1.06 |
| macromolecule catabolic process | 194 | 1.12 | -1.02 | 1.09 | 1.02 | 1.13 | 1.1 | 1.16 |
| biosynthetic process | 1003 | 1.19 | 1.05 | 1.09 | 1.06 | 1.14 | 1.18 | 1.04 |
| macromolecule biosynthetic process | 633 | 1.18 | 1.09 | 1.17 | 1.11 | 1.2 | 1.2 | 1.03 |
| aerobic respiration | 37 | -1.01 | 1.07 | 1.26 | -1.07 | 1.11 | 1.57 | -1.08 |
| anaerobic respiration | 1 | 1.08 | 1.05 | 1.26 | 1.07 | 1.31 | 1.33 | 1.1 |
| fatty acid catabolic process | 14 | -1.01 | 1.07 | -1.43 | 1.07 | 1.18 | -1.97 | -1.26 |
| cellular amino acid catabolic process | 26 | 1.02 | -1.11 | -1.05 | -1.1 | 1.01 | 1.7 | -1.18 |
| glutamine family amino acid metabolic process | 23 | -1.67 | -1.06 | -1.81 | -1.18 | 1.04 | -1.04 | -1.34 |
| glutamine family amino acid catabolic process | 9 | -1.31 | -1.36 | -1.54 | -1.44 | 1.15 | 1.66 | -1.26 |
| aspartate family amino acid metabolic process | 11 | 1.2 | -1.0 | -1.12 | 1.08 | -1.05 | 1.59 | -1.22 |
| aspartate family amino acid biosynthetic process | 4 | 1.12 | 1.16 | -1.0 | 1.25 | 1.08 | 1.34 | -1.11 |
| aspartate family amino acid catabolic process | 2 | -1.01 | -1.12 | -1.04 | 1.37 | -1.27 | 1.52 | -1.61 |
| serine family amino acid metabolic process | 17 | 1.58 | 1.09 | 1.09 | 1.0 | -1.11 | 1.65 | 1.1 |
| serine family amino acid biosynthetic process | 8 | 2.16 | 1.12 | -1.08 | 1.04 | -1.18 | 1.67 | 1.19 |
| serine family amino acid catabolic process | 6 | 1.21 | 1.07 | 1.39 | -1.04 | -1.02 | 1.86 | -1.02 |
| aromatic amino acid family metabolic process | 19 | 1.42 | -1.17 | -1.18 | -1.07 | -1.13 | 1.48 | 1.04 |
| aromatic amino acid family catabolic process | 6 | 1.4 | -1.16 | -1.06 | -1.08 | -1.08 | 1.95 | -1.14 |
| pyruvate family amino acid metabolic process | 1 | 1.7 | 1.2 | 1.13 | 1.08 | -1.16 | -1.08 | 1.29 |
| pyruvate family amino acid biosynthetic process | 1 | 1.7 | 1.2 | 1.13 | 1.08 | -1.16 | -1.08 | 1.29 |
| branched chain family amino acid metabolic process | 7 | 1.25 | 1.54 | 1.17 | 1.06 | -1.04 | 1.35 | 1.33 |
| branched chain family amino acid biosynthetic process | 1 | 5.21 | 1.63 | 1.44 | -1.45 | -2.46 | 1.42 | 2.6 |
| branched chain family amino acid catabolic process | 2 | 1.3 | 1.33 | 1.23 | 1.08 | 1.06 | 1.1 | -1.0 |
| glutamine family amino acid biosynthetic process | 13 | -1.5 | -1.15 | -1.86 | -1.29 | -1.1 | 1.05 | -1.0 |
| methionine biosynthetic process | 2 | 1.35 | 1.12 | -1.14 | 1.23 | 1.0 | 1.41 | -1.09 |
| methionine catabolic process | 1 | 1.48 | 1.28 | 1.32 | 1.05 | 1.07 | 1.16 | -1.01 |
| glycoprotein metabolic process | 69 | 1.16 | 1.07 | 1.11 | 1.04 | 1.13 | 1.16 | 1.31 |
| glycoprotein biosynthetic process | 66 | 1.17 | 1.06 | 1.1 | 1.04 | 1.12 | 1.16 | 1.31 |
| lipopolysaccharide biosynthetic process | 2 | 1.01 | -1.07 | -1.02 | 1.06 | 1.11 | -1.07 | -1.11 |
| lipoate metabolic process | 3 | 1.25 | 1.0 | 1.66 | -1.03 | -1.05 | 2.4 | -1.24 |
| coenzyme biosynthetic process | 38 | 1.65 | 1.09 | 1.07 | 1.05 | 1.11 | 1.24 | 1.16 |
| coenzyme catabolic process | 37 | -1.01 | 1.1 | 1.27 | -1.05 | 1.08 | 1.5 | -1.04 |
| vitamin biosynthetic process | 9 | 1.68 | 1.27 | -1.29 | -1.05 | -1.17 | 1.44 | 1.19 |
| nucleobase metabolic process | 14 | 1.45 | 1.02 | -1.06 | -1.01 | -1.04 | 1.16 | -1.01 |
| purine base biosynthetic process | 4 | 1.18 | 1.13 | 1.03 | 1.06 | -1.06 | 1.6 | -1.13 |
| nucleoside metabolic process | 33 | -1.08 | 1.04 | 1.04 | 1.11 | 1.12 | -1.17 | -1.05 |
| nucleotide metabolic process | 167 | 1.11 | 1.05 | 1.05 | 1.02 | 1.07 | 1.16 | 1.0 |
| ribonucleoside metabolic process | 22 | -1.04 | 1.14 | 1.02 | 1.2 | 1.26 | -1.16 | -1.09 |
| nucleoside monophosphate metabolic process | 51 | 1.01 | 1.08 | 1.01 | 1.01 | 1.01 | 1.24 | 1.04 |
| nucleoside monophosphate biosynthetic process | 48 | 1.01 | 1.08 | -1.0 | 1.01 | -1.0 | 1.23 | 1.02 |
| purine nucleoside monophosphate metabolic process | 16 | -1.24 | 1.08 | -1.05 | -1.03 | -1.12 | 1.38 | -1.02 |
| purine nucleoside monophosphate biosynthetic process | 16 | -1.24 | 1.08 | -1.05 | -1.03 | -1.12 | 1.38 | -1.02 |
| pyrimidine nucleoside monophosphate metabolic process | 6 | 1.07 | 1.17 | 1.04 | 1.26 | 1.2 | -1.05 | -1.08 |
| pyrimidine nucleoside monophosphate biosynthetic process | 6 | 1.07 | 1.17 | 1.04 | 1.26 | 1.2 | -1.05 | -1.08 |
| nucleoside diphosphate metabolic process | 9 | -1.16 | 1.05 | 1.27 | 1.28 | 1.4 | -1.29 | -1.31 |
| nucleoside diphosphate biosynthetic process | 3 | -1.09 | 1.03 | 1.21 | 1.32 | 1.42 | -1.7 | -1.39 |
| purine nucleoside diphosphate metabolic process | 2 | -1.29 | -1.01 | -1.04 | 1.37 | 1.42 | -2.05 | -1.57 |
| purine nucleoside diphosphate biosynthetic process | 2 | -1.29 | -1.01 | -1.04 | 1.37 | 1.42 | -2.05 | -1.57 |
| pyrimidine nucleoside diphosphate metabolic process | 1 | 1.27 | 1.13 | 1.91 | 1.21 | 1.41 | -1.17 | -1.08 |
| pyrimidine nucleoside diphosphate biosynthetic process | 1 | 1.27 | 1.13 | 1.91 | 1.21 | 1.41 | -1.17 | -1.08 |
| nucleoside triphosphate metabolic process | 78 | 1.23 | 1.01 | 1.2 | 1.0 | 1.11 | 1.29 | -1.05 |
| nucleoside triphosphate biosynthetic process | 29 | 1.28 | 1.05 | 1.25 | -1.01 | 1.13 | 1.56 | -1.1 |
| nucleoside triphosphate catabolic process | 39 | 1.16 | 1.04 | 1.14 | 1.09 | 1.12 | 1.08 | 1.01 |
| purine nucleoside triphosphate metabolic process | 77 | 1.24 | 1.01 | 1.2 | -1.0 | 1.1 | 1.31 | -1.04 |
| purine nucleoside triphosphate biosynthetic process | 29 | 1.28 | 1.05 | 1.25 | -1.01 | 1.13 | 1.56 | -1.1 |
| purine nucleoside triphosphate catabolic process | 39 | 1.16 | 1.04 | 1.14 | 1.09 | 1.12 | 1.08 | 1.01 |
| pyrimidine nucleoside triphosphate metabolic process | 5 | -1.06 | 1.02 | 1.3 | 1.3 | 1.44 | -1.22 | -1.3 |
| pyrimidine nucleoside triphosphate biosynthetic process | 4 | 1.0 | 1.09 | 1.36 | 1.22 | 1.36 | 1.06 | -1.14 |
| purine ribonucleotide metabolic process | 96 | 1.14 | 1.02 | 1.15 | 1.0 | 1.07 | 1.3 | -1.04 |
| purine ribonucleotide biosynthetic process | 48 | 1.07 | 1.06 | 1.13 | -1.0 | 1.06 | 1.42 | -1.08 |
| purine ribonucleotide catabolic process | 39 | 1.16 | 1.04 | 1.14 | 1.09 | 1.12 | 1.08 | 1.01 |
| ribonucleoside monophosphate biosynthetic process | 21 | -1.11 | 1.1 | -1.01 | 1.02 | -1.07 | 1.26 | -1.02 |
| deoxyribonucleoside monophosphate biosynthetic process | 2 | 1.05 | 1.03 | -1.1 | 1.26 | 1.31 | 1.33 | -1.09 |
| ribonucleoside monophosphate metabolic process | 21 | -1.11 | 1.1 | -1.01 | 1.02 | -1.07 | 1.26 | -1.02 |
| deoxyribonucleoside monophosphate metabolic process | 2 | 1.05 | 1.03 | -1.1 | 1.26 | 1.31 | 1.33 | -1.09 |
| nucleoside biosynthetic process | 7 | 1.07 | 1.09 | -1.03 | 1.13 | 1.19 | -1.05 | -1.12 |
| nucleoside catabolic process | 3 | -2.8 | 1.04 | -1.39 | 1.2 | 1.32 | -1.87 | -1.06 |
| nucleotide biosynthetic process | 88 | 1.12 | 1.07 | 1.09 | 1.01 | 1.06 | 1.32 | -1.01 |
| nucleotide catabolic process | 49 | -1.06 | 1.03 | 1.02 | 1.06 | 1.05 | -1.13 | 1.02 |
| purine ribonucleoside monophosphate metabolic process | 16 | -1.24 | 1.08 | -1.05 | -1.03 | -1.12 | 1.38 | -1.02 |
| purine ribonucleoside monophosphate biosynthetic process | 16 | -1.24 | 1.08 | -1.05 | -1.03 | -1.12 | 1.38 | -1.02 |
| pyrimidine ribonucleoside monophosphate metabolic process | 4 | 1.08 | 1.24 | 1.11 | 1.27 | 1.15 | -1.24 | -1.08 |
| pyrimidine ribonucleoside monophosphate biosynthetic process | 4 | 1.08 | 1.24 | 1.11 | 1.27 | 1.15 | -1.24 | -1.08 |
| pyrimidine deoxyribonucleoside monophosphate metabolic process | 2 | 1.05 | 1.03 | -1.1 | 1.26 | 1.31 | 1.33 | -1.09 |
| pyrimidine deoxyribonucleoside monophosphate biosynthetic process | 2 | 1.05 | 1.03 | -1.1 | 1.26 | 1.31 | 1.33 | -1.09 |
| purine ribonucleoside diphosphate metabolic process | 2 | -1.29 | -1.01 | -1.04 | 1.37 | 1.42 | -2.05 | -1.57 |
| purine ribonucleoside diphosphate biosynthetic process | 2 | -1.29 | -1.01 | -1.04 | 1.37 | 1.42 | -2.05 | -1.57 |
| ribonucleoside diphosphate metabolic process | 2 | -1.29 | -1.01 | -1.04 | 1.37 | 1.42 | -2.05 | -1.57 |
| deoxyribonucleoside diphosphate metabolic process | 2 | -1.56 | -1.01 | 1.45 | 1.31 | 1.53 | -1.54 | -1.56 |
| cyclic nucleotide metabolic process | 28 | 1.1 | 1.08 | 1.04 | -1.0 | 1.05 | 1.22 | 1.09 |
| muscle attachment | 28 | 1.6 | 1.08 | 1.3 | -1.01 | 1.35 | 1.36 | 1.25 |
| ribonucleoside diphosphate biosynthetic process | 2 | -1.29 | -1.01 | -1.04 | 1.37 | 1.42 | -2.05 | -1.57 |
| deoxyribonucleoside diphosphate biosynthetic process | 1 | 1.27 | 1.13 | 1.91 | 1.21 | 1.41 | -1.17 | -1.08 |
| cyclic nucleotide biosynthetic process | 25 | 1.1 | 1.06 | 1.01 | -1.01 | 1.03 | 1.2 | 1.06 |
| pyrimidine deoxyribonucleoside diphosphate metabolic process | 1 | 1.27 | 1.13 | 1.91 | 1.21 | 1.41 | -1.17 | -1.08 |
| pyrimidine deoxyribonucleoside diphosphate biosynthetic process | 1 | 1.27 | 1.13 | 1.91 | 1.21 | 1.41 | -1.17 | -1.08 |
| ribonucleoside triphosphate metabolic process | 77 | 1.24 | 1.01 | 1.2 | -1.0 | 1.1 | 1.31 | -1.04 |
| deoxyribonucleoside triphosphate metabolic process | 1 | -1.36 | -1.27 | 1.07 | 1.7 | 1.82 | -3.47 | -2.13 |
| ribonucleoside triphosphate biosynthetic process | 29 | 1.28 | 1.05 | 1.25 | -1.01 | 1.13 | 1.56 | -1.1 |
| ribonucleoside triphosphate catabolic process | 39 | 1.16 | 1.04 | 1.14 | 1.09 | 1.12 | 1.08 | 1.01 |
| purine ribonucleoside triphosphate metabolic process | 77 | 1.24 | 1.01 | 1.2 | -1.0 | 1.1 | 1.31 | -1.04 |
| purine ribonucleoside triphosphate biosynthetic process | 29 | 1.28 | 1.05 | 1.25 | -1.01 | 1.13 | 1.56 | -1.1 |
| purine ribonucleoside triphosphate catabolic process | 39 | 1.16 | 1.04 | 1.14 | 1.09 | 1.12 | 1.08 | 1.01 |
| pyrimidine ribonucleoside triphosphate metabolic process | 4 | 1.0 | 1.09 | 1.36 | 1.22 | 1.36 | 1.06 | -1.14 |
| pyrimidine ribonucleoside triphosphate biosynthetic process | 4 | 1.0 | 1.09 | 1.36 | 1.22 | 1.36 | 1.06 | -1.14 |
| pyrimidine deoxyribonucleoside triphosphate metabolic process | 1 | -1.36 | -1.27 | 1.07 | 1.7 | 1.82 | -3.47 | -2.13 |
| pyrimidine ribonucleotide metabolic process | 8 | 1.04 | 1.17 | 1.23 | 1.24 | 1.25 | -1.08 | -1.11 |
| pyrimidine deoxyribonucleotide metabolic process | 5 | 1.18 | -1.03 | 1.1 | 1.33 | 1.37 | -1.01 | -1.27 |
| pyrimidine ribonucleotide biosynthetic process | 8 | 1.04 | 1.17 | 1.23 | 1.24 | 1.25 | -1.08 | -1.11 |
| pyrimidine deoxyribonucleotide biosynthetic process | 3 | 1.12 | 1.06 | 1.16 | 1.24 | 1.35 | 1.15 | -1.08 |
| nucleotide-sugar metabolic process | 4 | -1.38 | 1.14 | 1.12 | 1.14 | 1.22 | -1.6 | -1.01 |
| nucleotide-sugar biosynthetic process | 4 | -1.38 | 1.14 | 1.12 | 1.14 | 1.22 | -1.6 | -1.01 |
| thiamine diphosphate biosynthetic process | 1 | 1.06 | 1.62 | -1.26 | 1.22 | -1.14 | -1.11 | 1.12 |
| riboflavin biosynthetic process | 2 | 1.78 | 2.05 | -2.11 | -2.16 | -2.52 | 2.14 | 1.44 |
| menaquinone metabolic process | 1 | 1.46 | 1.06 | 1.55 | -1.07 | 1.35 | 1.82 | 1.43 |
| menaquinone biosynthetic process | 1 | 1.46 | 1.06 | 1.55 | -1.07 | 1.35 | 1.82 | 1.43 |
| glycolipid biosynthetic process | 7 | 1.02 | 1.24 | 1.08 | -1.02 | 1.04 | -1.16 | 1.9 |
| glucan biosynthetic process | 5 | 2.74 | -1.19 | -1.42 | 1.04 | 1.07 | 2.78 | 1.29 |
| glucan catabolic process | 1 | 2.98 | -1.08 | 1.5 | -1.31 | 1.12 | 4.86 | 1.33 |
| peptidoglycan catabolic process | 12 | -1.13 | -1.64 | -1.19 | -1.04 | -1.82 | -1.69 | -1.29 |
| 10-formyltetrahydrofolate metabolic process | 2 | 2.31 | -1.37 | 1.2 | -1.19 | -1.05 | 2.92 | 1.06 |
| 10-formyltetrahydrofolate catabolic process | 1 | -1.35 | 1.22 | 1.7 | 1.18 | 1.15 | -1.09 | 1.03 |
| ribonucleotide metabolic process | 101 | 1.15 | 1.03 | 1.15 | 1.01 | 1.07 | 1.28 | -1.04 |
| ribonucleotide biosynthetic process | 53 | 1.09 | 1.07 | 1.13 | 1.01 | 1.06 | 1.37 | -1.08 |
| ribonucleotide catabolic process | 39 | 1.16 | 1.04 | 1.14 | 1.09 | 1.12 | 1.08 | 1.01 |
| deoxyribonucleotide metabolic process | 7 | -1.11 | 1.02 | 1.04 | 1.35 | 1.33 | -1.29 | -1.3 |
| deoxyribonucleotide biosynthetic process | 3 | 1.12 | 1.06 | 1.16 | 1.24 | 1.35 | 1.15 | -1.08 |
| deoxyribonucleotide catabolic process | 1 | -1.52 | 1.52 | -1.38 | 1.37 | -1.06 | -2.8 | 1.21 |
| 2'-deoxyribonucleotide biosynthetic process | 3 | 1.12 | 1.06 | 1.16 | 1.24 | 1.35 | 1.15 | -1.08 |
| response to temperature stimulus | 95 | 1.16 | -1.06 | 1.01 | -1.09 | 1.09 | 1.11 | 1.17 |
| cellular response to starvation | 14 | 1.29 | 1.12 | 1.14 | 1.05 | 1.06 | 1.68 | 1.18 |
| response to desiccation | 2 | 1.15 | 1.22 | 1.01 | 1.25 | -1.19 | 1.08 | -1.16 |
| response to humidity | 2 | 2.35 | -1.29 | -1.39 | -1.42 | -1.06 | 2.35 | 1.81 |
| cell outer membrane | 2 | 2.45 | -1.01 | 1.06 | 1.01 | 1.14 | 1.16 | 1.19 |
| bacterial-type flagellum | 1 | -1.07 | -1.0 | -1.08 | -1.04 | -1.09 | -1.14 | 1.31 |
| nucleoid | 1 | 1.18 | 1.16 | 1.75 | 1.38 | 1.68 | 1.06 | -1.24 |
| flagellum assembly | 10 | -1.03 | 1.09 | 1.07 | 1.09 | 1.11 | 1.01 | -1.02 |
| GDP-mannose biosynthetic process | 1 | -1.31 | 1.22 | 1.33 | 1.17 | 1.22 | -1.88 | 1.41 |
| snRNA transcription | 5 | 1.23 | 1.22 | 1.33 | 1.11 | 1.31 | 1.28 | 1.23 |
| rRNA transcription | 1 | -1.18 | 1.5 | 1.2 | 1.21 | 1.41 | -1.06 | 1.52 |
| tRNA transcription | 1 | 1.67 | 1.25 | 1.56 | 1.07 | 1.53 | 1.47 | 1.26 |
| protein secretion | 28 | 1.07 | 1.14 | 1.2 | 1.08 | 1.17 | 1.16 | 1.26 |
| amine metabolic process | 302 | -1.22 | -1.19 | -1.33 | -1.18 | -1.18 | -1.09 | 1.09 |
| amine biosynthetic process | 54 | 1.05 | -1.02 | -1.24 | -1.05 | -1.1 | 1.19 | 1.08 |
| amine catabolic process | 29 | -1.04 | -1.07 | -1.13 | -1.1 | -1.04 | 1.35 | -1.08 |
| oligosaccharide metabolic process | 23 | 1.34 | -1.09 | -1.13 | -1.14 | -1.02 | 1.21 | 1.32 |
| oligosaccharide biosynthetic process | 18 | 1.33 | -1.18 | -1.13 | -1.19 | -1.01 | 1.24 | 1.29 |
| response to radiation | 119 | 1.1 | -1.05 | 1.01 | -1.01 | 1.06 | 1.1 | 1.04 |
| response to drug | 6 | -1.83 | -1.23 | -1.66 | 1.04 | 1.16 | -1.94 | -1.32 |
| phenylalanine-tRNA ligase complex | 2 | -1.21 | 1.26 | 1.34 | 1.3 | 1.09 | -1.32 | -1.12 |
| glycerol-3-phosphate dehydrogenase complex | 7 | 1.04 | 1.0 | -1.12 | 1.1 | 1.28 | 1.07 | -1.09 |
| dihydrolipoyl dehydrogenase complex | 1 | -1.4 | -1.14 | 2.27 | -1.25 | -1.48 | 1.94 | -1.5 |
| pyruvate dehydrogenase complex | 4 | 1.04 | 1.05 | 1.45 | 1.06 | 1.07 | 1.89 | -1.15 |
| mitochondrial oxoglutarate dehydrogenase complex | 3 | 1.31 | 1.13 | 2.04 | -1.28 | 1.13 | 2.68 | -1.02 |
| oxoglutarate dehydrogenase complex | 3 | 1.31 | 1.13 | 2.04 | -1.28 | 1.13 | 2.68 | -1.02 |
| succinate-CoA ligase complex (ADP-forming) | 1 | 1.64 | -1.15 | 2.7 | -1.43 | 1.36 | 4.94 | -1.1 |
| endopeptidase Clp complex | 1 | -1.01 | 1.55 | 1.36 | 1.04 | 1.26 | 2.38 | 1.15 |
| peptidase activity, acting on L-amino acid peptides | 556 | -1.79 | -1.27 | -1.34 | -1.05 | -1.19 | -1.74 | -1.45 |
| 2'-deoxyribonucleotide metabolic process | 5 | 1.18 | -1.03 | 1.1 | 1.33 | 1.37 | -1.01 | -1.27 |
| phospholipid catabolic process | 4 | -7.22 | -4.62 | -6.48 | 1.55 | -1.18 | -4.98 | -5.49 |
| folic acid-containing compound biosynthetic process | 6 | 1.83 | -1.4 | -1.53 | -1.23 | -1.52 | 1.23 | 1.04 |
| folic acid-containing compound catabolic process | 1 | -1.35 | 1.22 | 1.7 | 1.18 | 1.15 | -1.09 | 1.03 |
| response to toxin | 23 | -1.02 | 1.14 | -1.39 | -1.1 | -1.24 | -1.6 | 1.25 |
| toxin metabolic process | 10 | 1.1 | -1.02 | -1.79 | -1.06 | -1.28 | -1.46 | 1.06 |
| pathogenesis | 1 | -117.62 | -1.03 | 2.87 | -1.15 | -1.31 | -23.49 | -2.28 |
| viral reproduction | 4 | -1.08 | 1.06 | 1.14 | -1.14 | -1.13 | 1.44 | 1.18 |
| toxin catabolic process | 4 | -1.27 | -1.2 | -4.97 | -1.06 | -1.84 | -3.14 | 1.18 |
| response to heat | 73 | 1.17 | -1.05 | 1.01 | -1.1 | 1.11 | 1.13 | 1.19 |
| response to cold | 2 | 1.0 | 1.06 | -1.05 | 1.0 | -1.03 | -1.07 | -1.17 |
| response to xenobiotic stimulus | 10 | 1.1 | -1.02 | -1.79 | -1.06 | -1.28 | -1.46 | 1.06 |
| response to UV | 20 | -1.19 | -1.26 | -1.33 | -1.19 | -1.15 | 1.11 | -1.01 |
| response to metal ion | 16 | -1.69 | 1.12 | 1.25 | 1.08 | 1.07 | -1.25 | -1.13 |
| response to water deprivation | 3 | -1.08 | -2.2 | -2.66 | -2.3 | -1.91 | 1.61 | -1.02 |
| response to water | 7 | 1.12 | -1.5 | -1.68 | -1.54 | -1.43 | 1.5 | 1.17 |
| response to light stimulus | 106 | 1.09 | -1.08 | -1.02 | -1.02 | 1.04 | 1.12 | 1.04 |
| microtubule-based flagellum | 2 | -1.07 | 1.15 | -1.03 | 1.08 | 1.03 | -1.0 | 1.04 |
| NAD biosynthetic process | 3 | 1.9 | 1.5 | -1.09 | 1.01 | 1.09 | 1.23 | 1.86 |
| glyoxylate catabolic process | 1 | 1.1 | -1.81 | -2.08 | -1.81 | -1.6 | 3.21 | -1.83 |
| carnitine metabolic process | 2 | -1.01 | -1.08 | -1.06 | -1.12 | -1.16 | 1.15 | -1.15 |
| gamma-aminobutyric acid metabolic process | 3 | -1.67 | 1.65 | -1.0 | -1.05 | -1.15 | -1.16 | -1.07 |
| gamma-aminobutyric acid biosynthetic process | 1 | -1.13 | 1.06 | -1.08 | -1.01 | -1.16 | -1.21 | -1.16 |
| gamma-aminobutyric acid catabolic process | 1 | -2.19 | 1.37 | 2.06 | 1.34 | 1.0 | 1.58 | -1.11 |
| RNA modification | 40 | 1.41 | 1.21 | 1.2 | 1.19 | 1.24 | 1.25 | 1.03 |
| RNA capping | 3 | 1.59 | 1.23 | 1.42 | 1.29 | 1.28 | 1.33 | 1.13 |
| energy taxis | 14 | 1.37 | -1.07 | 1.01 | -1.18 | -1.0 | 1.35 | 1.36 |
| electron transporter, transferring electrons within CoQH2-cytochrome c reductase complex activity | 2 | -1.21 | 1.01 | 1.07 | -1.08 | -1.12 | 1.17 | -1.37 |
| electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity | 2 | 1.19 | 1.13 | 1.42 | -1.04 | 1.3 | 1.64 | -1.09 |
| glutathione disulfide oxidoreductase activity | 5 | 1.08 | 1.22 | -1.06 | 1.06 | 1.17 | -1.15 | 1.05 |
| fertilization | 22 | 1.04 | 1.13 | 1.13 | 1.08 | 1.1 | 1.04 | 1.03 |
| detection of external stimulus | 59 | 1.07 | -1.17 | -1.01 | 1.04 | 1.0 | -1.01 | -1.14 |
| detection of abiotic stimulus | 53 | 1.09 | -1.1 | -1.03 | 1.04 | 1.06 | 1.03 | -1.05 |
| detection of light stimulus | 48 | 1.08 | -1.11 | -1.03 | 1.04 | 1.06 | 1.01 | -1.07 |
| detection of visible light | 24 | 1.14 | -1.06 | -1.0 | 1.09 | 1.08 | 1.04 | -1.06 |
| rhodopsin mediated phototransduction | 15 | 1.23 | 1.02 | 1.1 | 1.07 | 1.08 | 1.05 | 1.02 |
| UV-A, blue light phototransduction | 2 | 1.02 | -1.04 | -1.16 | 1.02 | -1.2 | 1.38 | -1.05 |
| detection of UV | 5 | -1.46 | -1.43 | -1.51 | 1.22 | -1.06 | -1.54 | -1.62 |
| detection of chemical stimulus | 84 | -1.02 | 1.01 | -1.0 | 1.01 | -1.0 | 1.03 | -1.06 |
| detection of nutrient | 1 | 1.24 | 1.34 | 2.02 | 1.2 | 1.78 | 5.29 | 1.65 |
| detection of biotic stimulus | 6 | -1.12 | -1.97 | 1.21 | 1.05 | -1.56 | -1.46 | -2.39 |
| detection of virus | 1 | -1.09 | 1.04 | 3.03 | -1.09 | 1.76 | 1.29 | 1.77 |
| detection of symbiont | 1 | -1.09 | 1.04 | 3.03 | -1.09 | 1.76 | 1.29 | 1.77 |
| response to external stimulus | 341 | 1.09 | -1.0 | 1.13 | -1.01 | 1.17 | 1.21 | 1.17 |
| response to biotic stimulus | 155 | 1.24 | -1.24 | -1.18 | -1.24 | -1.09 | 1.12 | 1.16 |
| response to symbiont | 7 | -1.49 | 1.04 | -1.1 | 1.15 | 1.18 | -1.6 | -1.03 |
| response to symbiotic bacterium | 2 | -5.67 | -1.13 | -2.52 | 1.36 | 1.12 | -4.93 | -1.8 |
| response to wounding | 31 | 1.1 | -1.06 | 1.05 | -1.14 | 1.05 | 1.17 | 1.32 |
| response to other organism | 153 | 1.25 | -1.25 | -1.19 | -1.24 | -1.1 | 1.12 | 1.16 |
| response to bacterium | 100 | 1.24 | -1.29 | -1.25 | -1.25 | -1.11 | 1.14 | 1.1 |
| response to fungus | 31 | 1.98 | -1.28 | -1.31 | -1.33 | -1.26 | 1.23 | 1.29 |
| response to insect | 1 | 23.01 | -3.14 | -2.9 | -3.17 | -2.48 | 1.73 | 2.58 |
| response to abiotic stimulus | 224 | 1.14 | -1.03 | 1.04 | -1.02 | 1.09 | 1.14 | 1.1 |
| response to gravity | 12 | 1.09 | -1.01 | 1.03 | 1.04 | 1.1 | 1.2 | -1.03 |
| response to blue light | 1 | 1.37 | -1.11 | -1.04 | 1.15 | 1.03 | 2.71 | 1.13 |
| response to red or far red light | 2 | 1.78 | 1.07 | -1.24 | 1.28 | -1.04 | 1.5 | -1.03 |
| response to light intensity | 3 | 2.14 | -1.51 | 1.43 | -1.06 | -1.34 | 1.03 | -1.36 |
| response to high light intensity | 1 | 3.41 | -1.85 | -1.6 | -1.39 | -1.6 | 6.88 | -1.28 |
| photoperiodism | 7 | 1.11 | 1.14 | 1.05 | 1.16 | 1.0 | 1.28 | 1.14 |
| entrainment of circadian clock | 12 | 1.23 | 1.12 | 1.14 | 1.12 | 1.09 | 1.34 | 1.18 |
| UV protection | 1 | 1.47 | 1.08 | -1.62 | 1.31 | -1.48 | 3.54 | -1.09 |
| response to salt stress | 7 | 1.27 | -1.08 | 1.33 | -1.12 | 1.11 | 1.26 | 1.47 |
| catechol-containing compound metabolic process | 9 | -1.02 | -1.18 | -1.26 | -1.16 | -1.28 | -1.1 | 1.52 |
| catechol-containing compound biosynthetic process | 3 | -1.04 | -2.12 | -1.47 | -1.4 | -1.53 | 1.41 | 2.4 |
| response to endogenous stimulus | 81 | 1.15 | 1.05 | 1.35 | 1.05 | 1.23 | 1.3 | 1.17 |
| response to hormone stimulus | 64 | 1.07 | 1.08 | 1.26 | 1.05 | 1.23 | 1.29 | 1.21 |
| detection of carbohydrate stimulus | 1 | -1.49 | -1.13 | 1.17 | 1.11 | 1.02 | -1.25 | -1.24 |
| detection of sucrose stimulus | 1 | -1.49 | -1.13 | 1.17 | 1.11 | 1.02 | -1.25 | -1.24 |
| detection of hexose stimulus | 1 | -1.49 | -1.13 | 1.17 | 1.11 | 1.02 | -1.25 | -1.24 |
| response to carbohydrate stimulus | 6 | 1.99 | -1.0 | 1.23 | 1.06 | 1.32 | 1.59 | 1.29 |
| response to sucrose stimulus | 3 | 2.79 | -1.08 | 1.21 | 1.01 | 1.44 | 1.96 | 1.48 |
| response to hexose stimulus | 2 | -1.1 | 1.06 | 1.16 | 1.15 | 1.11 | -1.04 | -1.08 |
| response to glucose stimulus | 2 | -1.1 | 1.06 | 1.16 | 1.15 | 1.11 | -1.04 | -1.08 |
| hormone-mediated signaling pathway | 22 | 1.14 | 1.08 | 1.04 | 1.01 | 1.16 | 1.17 | 1.17 |
| carbohydrate utilization | 1 | 1.08 | -1.16 | -1.09 | 1.12 | -1.12 | 1.04 | -1.07 |
| regulation of asymmetric cell division | 2 | -1.7 | 1.32 | 1.22 | 1.23 | 1.24 | -1.9 | -1.09 |
| post-embryonic development | 443 | 1.18 | 1.05 | 1.22 | 1.02 | 1.25 | 1.26 | 1.22 |
| embryo development ending in birth or egg hatching | 163 | 1.1 | 1.06 | 1.3 | 1.03 | 1.33 | 1.32 | 1.23 |
| regulation of mitotic cell cycle, embryonic | 8 | -1.01 | 1.14 | 1.15 | 1.12 | 1.19 | 1.01 | 1.02 |
| axis specification | 164 | 1.24 | 1.06 | 1.24 | 1.03 | 1.22 | 1.3 | 1.23 |
| specification of symmetry | 5 | -2.92 | 1.25 | 1.15 | -1.01 | 1.23 | -1.58 | 1.16 |
| unidimensional cell growth | 17 | 1.27 | 1.19 | 1.53 | 1.1 | 1.7 | 1.49 | 1.46 |
| SCF ubiquitin ligase complex | 14 | 1.41 | 1.21 | 1.25 | 1.2 | 1.31 | 1.18 | 1.12 |
| determination of bilateral symmetry | 5 | -2.92 | 1.25 | 1.15 | -1.01 | 1.23 | -1.58 | 1.16 |
| photoreceptor activity | 10 | -1.25 | -1.25 | -1.24 | -1.21 | -1.25 | 1.46 | -1.17 |
| blue light photoreceptor activity | 1 | 1.37 | -1.11 | -1.04 | 1.15 | 1.03 | 2.71 | 1.13 |
| post-embryonic morphogenesis | 362 | 1.17 | 1.06 | 1.22 | 1.03 | 1.28 | 1.25 | 1.24 |
| regulation of biosynthetic process | 771 | 1.15 | 1.07 | 1.16 | 1.05 | 1.17 | 1.16 | 1.13 |
| negative regulation of biosynthetic process | 219 | 1.16 | 1.07 | 1.18 | 1.04 | 1.23 | 1.2 | 1.14 |
| positive regulation of biosynthetic process | 252 | 1.15 | 1.03 | 1.12 | 1.02 | 1.13 | 1.2 | 1.17 |
| negative regulation of metabolic process | 330 | 1.18 | 1.06 | 1.17 | 1.05 | 1.2 | 1.2 | 1.12 |
| positive regulation of metabolic process | 281 | 1.17 | 1.04 | 1.14 | 1.02 | 1.14 | 1.22 | 1.17 |
| regulation of catabolic process | 74 | 1.15 | 1.14 | 1.25 | 1.08 | 1.23 | 1.24 | 1.17 |
| negative regulation of catabolic process | 8 | -1.33 | 1.25 | 1.01 | 1.18 | 1.01 | -1.53 | -1.03 |
| positive regulation of catabolic process | 7 | 1.19 | 1.14 | 1.23 | 1.17 | 1.1 | 1.13 | 1.03 |
| external side of plasma membrane | 4 | -1.07 | 1.05 | -1.01 | -1.05 | -1.13 | -1.02 | -1.0 |
| internal side of plasma membrane | 18 | 1.39 | 1.11 | 1.42 | 1.06 | 1.22 | 1.03 | 1.1 |
| epidermal cell differentiation | 43 | 1.39 | 1.06 | 1.43 | -1.01 | 1.34 | 1.35 | 1.33 |
| fatty acid elongase activity | 3 | 1.46 | -2.22 | -1.95 | -2.84 | -2.14 | -1.24 | 3.42 |
| basal plasma membrane | 1 | 1.33 | 1.32 | 2.26 | 1.11 | 1.52 | 1.39 | 1.48 |
| calcium-dependent protein serine/threonine kinase activity | 3 | 1.12 | 1.04 | 2.13 | -1.07 | 1.01 | 3.21 | -1.04 |
| proximal/distal axis specification | 1 | -1.27 | 1.59 | 1.07 | 1.18 | 1.42 | 1.16 | 1.47 |
| anterior/posterior axis specification | 120 | 1.19 | 1.06 | 1.26 | 1.05 | 1.25 | 1.26 | 1.2 |
| polarity specification of anterior/posterior axis | 5 | 1.18 | 1.04 | 1.06 | 1.17 | 1.12 | -1.03 | 1.06 |
| dorsal/ventral axis specification | 58 | 1.48 | 1.04 | 1.12 | 1.02 | 1.11 | 1.41 | 1.25 |
| polarity specification of dorsal/ventral axis | 5 | 1.18 | 1.04 | 1.06 | 1.17 | 1.12 | -1.03 | 1.06 |
| anterior/posterior pattern specification | 147 | 1.2 | 1.04 | 1.2 | 1.01 | 1.24 | 1.3 | 1.21 |
| dorsal/ventral pattern formation | 112 | 1.35 | 1.04 | 1.11 | 1.02 | 1.18 | 1.37 | 1.26 |
| proximal/distal pattern formation | 25 | 1.33 | 1.03 | 1.18 | 1.09 | 1.18 | 1.11 | 1.2 |
| regulation of signal transduction | 371 | 1.15 | 1.12 | 1.29 | 1.06 | 1.25 | 1.28 | 1.21 |
| positive regulation of signal transduction | 83 | 1.15 | 1.11 | 1.23 | 1.05 | 1.27 | 1.18 | 1.24 |
| negative regulation of signal transduction | 144 | 1.21 | 1.1 | 1.32 | 1.06 | 1.28 | 1.32 | 1.24 |
| cytidine deamination | 3 | -2.8 | 1.04 | -1.39 | 1.2 | 1.32 | -1.87 | -1.06 |
| cyclase activity | 29 | 1.15 | 1.04 | -1.01 | 1.0 | 1.03 | 1.12 | 1.07 |
| pseudouridine synthase activity | 10 | 1.65 | 1.34 | 1.08 | 1.11 | 1.11 | 1.56 | 1.14 |
| cell surface | 19 | -1.26 | 1.06 | 1.15 | 1.03 | 1.17 | 1.1 | 1.16 |
| cellular process | 5323 | 1.1 | 1.04 | 1.1 | 1.04 | 1.13 | 1.14 | 1.05 |
| response to extracellular stimulus | 43 | 1.1 | -1.0 | 1.12 | -1.02 | 1.16 | 1.24 | 1.19 |
| oocyte differentiation | 113 | 1.14 | 1.14 | 1.32 | 1.08 | 1.26 | 1.35 | 1.23 |
| negative regulation of cell fate specification | 18 | 1.02 | 1.09 | 1.22 | 1.03 | 1.24 | 1.12 | 1.22 |
| negative regulation of cardioblast cell fate specification | 7 | -1.04 | 1.07 | 1.14 | 1.01 | 1.15 | -1.01 | 1.35 |
| glial cell differentiation | 26 | 2.31 | -1.1 | 1.3 | -1.1 | 1.28 | 1.79 | 1.37 |
| cardioblast differentiation | 12 | 1.8 | -1.05 | 1.29 | -1.1 | 1.12 | 1.16 | 1.3 |
| gastrulation involving germ band extension | 30 | 1.11 | -1.0 | 1.2 | 1.03 | 1.32 | 1.16 | 1.06 |
| endosome membrane | 1 | 1.78 | 1.14 | -1.44 | 1.08 | 1.0 | 3.89 | 1.83 |
| meiotic chromosome condensation | 1 | -1.65 | -1.11 | -1.2 | -1.08 | 1.07 | -1.43 | -1.21 |
| response to organic substance | 192 | 1.24 | 1.01 | 1.14 | -1.04 | 1.16 | 1.25 | 1.21 |
| response to inorganic substance | 34 | -1.24 | 1.1 | 1.16 | 1.08 | 1.11 | -1.04 | -1.11 |
| response to carbon dioxide | 4 | -1.02 | 1.01 | 1.24 | -1.05 | 1.06 | 1.13 | -1.05 |
| response to zinc ion | 2 | -1.79 | 1.33 | -1.11 | -1.01 | -1.24 | -1.87 | -1.3 |
| specification of organ identity | 3 | 1.44 | -1.09 | -1.02 | 1.06 | -1.33 | -1.11 | 1.02 |
| cellular response to iron ion starvation | 1 | 1.19 | -1.04 | -1.01 | -1.09 | -1.15 | -1.15 | -1.12 |
| response to red light | 2 | 1.78 | 1.07 | -1.24 | 1.28 | -1.04 | 1.5 | -1.03 |
| regulation of proton transport | 1 | -1.15 | -1.06 | -1.02 | -1.13 | -1.25 | -1.1 | -1.2 |
| formation of organ boundary | 33 | 1.16 | 1.04 | 1.21 | 1.08 | 1.36 | 1.13 | 1.19 |
| response to X-ray | 4 | 1.1 | 1.11 | 1.34 | 1.23 | 1.84 | -1.18 | -1.06 |
| body morphogenesis | 19 | -1.01 | -1.0 | -1.07 | -1.04 | -1.14 | -1.01 | -1.14 |
| production of miRNAs involved in gene silencing by miRNA | 6 | 1.56 | 1.12 | 1.48 | 1.03 | 1.19 | 1.57 | 1.34 |
| response to ionizing radiation | 11 | 1.27 | 1.24 | 1.27 | 1.2 | 1.48 | -1.03 | 1.06 |
| response to lithium ion | 1 | -1.01 | -1.09 | -1.11 | -1.17 | -1.13 | 13.69 | -1.18 |
| response to organic nitrogen | 4 | 1.59 | -1.01 | 1.38 | 1.29 | 1.34 | 1.2 | -1.18 |
| endomembrane system organization | 2 | -1.26 | 1.07 | 1.65 | 1.14 | 1.45 | 1.05 | 1.02 |
| NADH dehydrogenase complex assembly | 1 | 1.08 | 1.05 | 1.31 | 1.03 | 1.07 | 1.99 | -1.52 |
| multicellular organismal aging | 114 | 1.25 | 1.12 | 1.11 | -1.02 | 1.16 | 1.37 | 1.25 |
| acireductone dioxygenase [iron(II)-requiring] activity | 1 | 1.79 | 1.32 | -1.5 | 1.17 | -1.1 | 1.63 | 1.03 |
| phosphatidylinositol-5-phosphate binding | 1 | -1.49 | -1.24 | 1.29 | 1.46 | 1.49 | -1.4 | 1.3 |
| membrane invagination | 183 | 1.09 | 1.08 | 1.18 | 1.07 | 1.21 | 1.18 | 1.13 |
| response to gamma radiation | 1 | 4.35 | 1.1 | 2.37 | 1.09 | 1.77 | 2.6 | 1.18 |
| carboxyl-O-methyltransferase activity | 2 | 1.17 | 1.1 | 1.15 | 1.29 | 1.38 | 1.07 | -1.31 |
| response to trehalose stimulus | 3 | 3.3 | 1.01 | 1.16 | -1.06 | 1.46 | 2.79 | 1.63 |
| chromocenter | 17 | 1.14 | 1.05 | 1.1 | 1.07 | 1.29 | 1.12 | 1.07 |
| temperature compensation of the circadian clock | 3 | -1.23 | 1.31 | 1.47 | 1.09 | 1.08 | -1.19 | 1.12 |
| double-stranded methylated DNA binding | 1 | -1.16 | -1.01 | -1.17 | -1.19 | -1.26 | -1.13 | -1.25 |
| regulation of G2/M transition of mitotic cell cycle | 72 | 1.2 | 1.14 | 1.34 | 1.2 | 1.4 | 1.22 | 1.12 |
| polyprenyldihydroxybenzoate methyltransferase activity | 1 | -1.1 | 1.09 | 1.22 | 1.29 | 1.3 | 1.24 | -1.16 |
| carotenoid dioxygenase activity | 1 | 1.12 | -1.01 | 1.14 | 1.06 | 1.09 | 1.14 | -1.06 |
| histone H3-K36 methylation | 1 | 1.08 | 1.46 | 1.5 | -1.1 | 1.11 | 1.44 | 1.39 |
| regulation of cell fate commitment | 26 | 1.1 | 1.06 | 1.12 | 1.01 | 1.18 | -1.03 | 1.12 |
| negative regulation of cell fate commitment | 18 | 1.02 | 1.09 | 1.22 | 1.03 | 1.24 | 1.12 | 1.22 |
| centriole-centriole cohesion | 1 | 3.23 | -1.13 | 1.38 | 1.13 | 1.64 | 2.38 | 1.04 |
| light-activated ion channel activity | 2 | -1.34 | -1.15 | -1.19 | 1.14 | -1.2 | -1.27 | -1.19 |
| negative regulation of peptidase activity | 3 | -1.25 | -1.08 | 1.67 | 1.12 | 1.52 | -1.68 | -1.23 |
| gene expression | 762 | 1.22 | 1.11 | 1.2 | 1.16 | 1.23 | 1.2 | 1.03 |
| regulation of gene expression | 882 | 1.18 | 1.08 | 1.18 | 1.07 | 1.18 | 1.19 | 1.12 |
| regulation of receptor activity | 4 | 1.16 | 1.09 | 1.49 | 1.02 | 1.37 | 1.48 | 1.2 |
| H3 histone acetyltransferase activity | 2 | -1.3 | 1.39 | 1.24 | 1.21 | 1.14 | 1.24 | 1.37 |
| H4 histone acetyltransferase activity | 4 | 1.11 | 1.19 | 1.15 | 1.22 | 1.15 | 1.12 | 1.31 |
| cytoplasmic stress granule | 1 | 1.54 | 1.07 | -1.03 | 1.11 | 1.17 | 1.09 | 1.04 |
| intercellular transport | 2 | 1.93 | -1.45 | -1.44 | -1.31 | -1.47 | 2.84 | 3.08 |
| proteasomal protein catabolic process | 20 | 1.05 | 1.14 | 1.1 | 1.07 | 1.12 | -1.0 | 1.14 |
| regulation of autophagy | 16 | 1.32 | 1.1 | 1.02 | 1.05 | 1.1 | 1.13 | 1.1 |
| negative regulation of autophagy | 4 | -1.64 | 1.35 | -1.16 | 1.08 | -1.15 | -1.93 | 1.04 |
| positive regulation of autophagy | 2 | -1.18 | 1.26 | 1.3 | 1.29 | 1.13 | -1.31 | -1.09 |
| regulation of phospholipase activity | 8 | 1.04 | 1.1 | 1.09 | 1.07 | 1.01 | 1.07 | 1.0 |
| positive regulation of phospholipase activity | 6 | 1.13 | 1.12 | 1.17 | 1.04 | 1.09 | 1.14 | 1.05 |
| negative regulation of phospholipase activity | 2 | -1.21 | 1.05 | -1.16 | 1.19 | -1.24 | -1.13 | -1.15 |
| regulation of calcium ion transport into cytosol | 1 | -1.3 | 1.21 | 1.06 | 1.22 | 1.05 | 1.22 | 1.37 |
| regulation of transposition, RNA-mediated | 2 | 1.32 | 1.05 | -1.25 | 1.18 | -1.12 | 1.33 | 1.08 |
| negative regulation of transposition, RNA-mediated | 2 | 1.32 | 1.05 | -1.25 | 1.18 | -1.12 | 1.33 | 1.08 |
| regulation of transposition | 5 | 1.07 | 1.14 | -1.03 | 1.17 | 1.03 | 1.08 | 1.04 |
| negative regulation of transposition | 5 | 1.07 | 1.14 | -1.03 | 1.17 | 1.03 | 1.08 | 1.04 |
| regulation of macromolecule biosynthetic process | 732 | 1.15 | 1.08 | 1.17 | 1.06 | 1.17 | 1.17 | 1.13 |
| positive regulation of macromolecule biosynthetic process | 215 | 1.13 | 1.05 | 1.14 | 1.04 | 1.15 | 1.2 | 1.16 |
| negative regulation of macromolecule biosynthetic process | 214 | 1.18 | 1.08 | 1.18 | 1.04 | 1.23 | 1.23 | 1.17 |
| regulation of glycoprotein biosynthetic process | 1 | -1.37 | -1.13 | -1.19 | -1.54 | 1.82 | 7.35 | 2.8 |
| positive regulation of phosphorus metabolic process | 8 | 1.51 | 1.21 | 1.82 | 1.05 | 1.45 | 2.01 | 1.56 |
| negative regulation of phosphorus metabolic process | 8 | 1.4 | 1.03 | 1.23 | 1.05 | 1.2 | 1.33 | 1.24 |
| regulation of cell cycle process | 162 | 1.23 | 1.12 | 1.27 | 1.13 | 1.35 | 1.28 | 1.15 |
| regulation of cellular ketone metabolic process | 2 | -1.15 | 1.19 | 1.31 | -1.0 | 1.1 | 1.01 | 1.21 |
| metalloenzyme regulator activity | 1 | 8.96 | -2.17 | 1.12 | -1.71 | -1.16 | 3.1 | 2.84 |
| regulation of adenylate cyclase activity involved in G-protein signaling pathway | 3 | -1.05 | 1.08 | -1.04 | 1.0 | -1.08 | -1.13 | -1.16 |
| positive regulation of adenylate cyclase activity by G-protein signaling pathway | 3 | -1.05 | 1.08 | -1.04 | 1.0 | -1.08 | -1.13 | -1.16 |
| miRNA metabolic process | 2 | 3.15 | 1.1 | 1.2 | 1.08 | -1.05 | 1.81 | 1.29 |
| regulation of lamellipodium assembly | 7 | -1.2 | 1.11 | 1.6 | 1.12 | 1.38 | -1.25 | -1.04 |
| negative regulation of lamellipodium assembly | 1 | -1.18 | 1.31 | 1.55 | 1.08 | 1.49 | 1.22 | 1.14 |
| regulation of cytoplasmic mRNA processing body assembly | 4 | 1.22 | 1.35 | 1.4 | 1.26 | 1.29 | 1.4 | 1.26 |
| positive regulation of macromolecule metabolic process | 246 | 1.12 | 1.06 | 1.17 | 1.04 | 1.15 | 1.2 | 1.17 |
| negative regulation of macromolecule metabolic process | 317 | 1.21 | 1.06 | 1.17 | 1.04 | 1.21 | 1.24 | 1.14 |
| positive regulation of cytoplasmic mRNA processing body assembly | 1 | 3.49 | 1.22 | 2.63 | -1.09 | 2.31 | 4.69 | 1.68 |
| posttranscriptional regulation of gene expression | 115 | 1.33 | 1.09 | 1.24 | 1.07 | 1.27 | 1.32 | 1.17 |
| developmental programmed cell death | 22 | 1.05 | 1.15 | 1.23 | 1.16 | 1.43 | 1.1 | 1.06 |
| regulation of intracellular protein kinase cascade | 60 | 1.06 | 1.18 | 1.27 | 1.1 | 1.28 | 1.26 | 1.23 |
| positive regulation of gene expression | 207 | 1.12 | 1.04 | 1.14 | 1.04 | 1.14 | 1.2 | 1.15 |
| negative regulation of gene expression | 260 | 1.2 | 1.06 | 1.15 | 1.04 | 1.2 | 1.23 | 1.14 |
| epithelial cell migration | 28 | 1.66 | -1.03 | 1.29 | -1.07 | 1.21 | 1.38 | 1.11 |
| regulation of epithelial cell migration | 5 | 2.13 | -1.29 | -1.2 | -1.27 | 1.01 | -1.13 | 2.14 |
| positive regulation of organelle organization | 20 | 1.05 | 1.11 | 1.26 | 1.08 | 1.36 | 1.03 | 1.17 |
| negative regulation of organelle organization | 35 | 1.05 | 1.13 | 1.38 | 1.11 | 1.41 | 1.3 | 1.13 |
| regulation of cell communication | 252 | 1.17 | 1.1 | 1.28 | 1.06 | 1.25 | 1.25 | 1.23 |
| positive regulation of cell communication | 85 | 1.13 | 1.11 | 1.24 | 1.05 | 1.27 | 1.19 | 1.24 |
| negative regulation of cell communication | 150 | 1.2 | 1.1 | 1.31 | 1.06 | 1.28 | 1.31 | 1.25 |
| epithelial structure maintenance | 13 | 1.3 | -1.11 | 1.06 | -1.15 | 1.26 | 1.38 | 1.31 |
| negative regulation of reactive oxygen species metabolic process | 1 | 1.06 | 1.25 | -1.01 | 1.13 | 1.01 | 1.03 | -1.27 |
| regulation of cellular carbohydrate metabolic process | 3 | -1.56 | -1.05 | -1.43 | -1.32 | 1.05 | 1.6 | 2.11 |
| positive regulation of cell development | 6 | 1.51 | 1.15 | 1.49 | 1.06 | 1.33 | 1.5 | 1.37 |
| negative regulation of cell development | 24 | -1.03 | 1.15 | 1.31 | 1.07 | 1.41 | 1.21 | 1.2 |
| positive regulation of intracellular protein kinase cascade | 18 | 1.04 | 1.19 | 1.27 | 1.03 | 1.17 | 1.26 | 1.3 |
| negative regulation of intracellular protein kinase cascade | 22 | 1.11 | 1.14 | 1.33 | 1.09 | 1.28 | 1.12 | 1.13 |
| positive regulation of sodium ion transport | 5 | 1.02 | 1.03 | 1.03 | 1.0 | -1.03 | 1.03 | -1.06 |
| regulation of cell morphogenesis involved in differentiation | 44 | 1.03 | 1.15 | 1.39 | 1.08 | 1.37 | 1.26 | 1.18 |
| meiotic DNA double-strand break formation involved in reciprocal meiotic recombination | 1 | 1.2 | 1.16 | 1.39 | 1.21 | 1.2 | 1.13 | -1.2 |
| regulation of multivesicular body size | 3 | 1.32 | -1.02 | 1.33 | 1.3 | 1.34 | 1.03 | -1.24 |
| regulation of multivesicular body size involved in endosome transport | 3 | 1.32 | -1.02 | 1.33 | 1.3 | 1.34 | 1.03 | -1.24 |
| regulation of peptidyl-threonine phosphorylation | 1 | 1.11 | 1.2 | 3.72 | 1.08 | 1.65 | 2.06 | 2.0 |
| positive regulation of peptidyl-threonine phosphorylation | 1 | 1.11 | 1.2 | 3.72 | 1.08 | 1.65 | 2.06 | 2.0 |
| regulation of cell-substrate adhesion | 2 | 1.91 | 1.0 | 2.08 | 1.07 | 1.96 | 1.63 | 1.31 |
| negative regulation of cell-substrate adhesion | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| regulation of hormone levels | 36 | 1.14 | -1.24 | -1.31 | -1.23 | -1.11 | -1.09 | 1.2 |
| regulation of mitochondrion organization | 6 | 1.18 | 1.14 | 1.11 | 1.16 | 1.06 | -1.11 | -1.04 |
| positive regulation of mitochondrion organization | 2 | -1.0 | 1.14 | -1.27 | 1.12 | -1.13 | -1.52 | -1.35 |
| regulation of centrosome duplication | 4 | 1.33 | 1.11 | 1.56 | 1.02 | 1.41 | 1.58 | 1.14 |
| transcription regulatory region DNA binding | 73 | 1.02 | 1.03 | 1.06 | 1.02 | 1.11 | 1.14 | 1.07 |
| regulation of chromatin assembly | 3 | 1.21 | 1.11 | 1.43 | 1.11 | 1.52 | 1.35 | 1.11 |
| cyclase regulator activity | 1 | 4.81 | -1.03 | -1.02 | 1.21 | 1.81 | 1.05 | 1.23 |
| cyclase activator activity | 1 | 4.81 | -1.03 | -1.02 | 1.21 | 1.81 | 1.05 | 1.23 |
| calcium-dependent protein kinase activity | 3 | 1.12 | 1.04 | 2.13 | -1.07 | 1.01 | 3.21 | -1.04 |
| endopeptidase regulator activity | 75 | 1.41 | -1.46 | -1.42 | -1.47 | -1.28 | 1.49 | 1.04 |
| positive regulation of phospholipase C activity | 6 | 1.13 | 1.12 | 1.17 | 1.04 | 1.09 | 1.14 | 1.05 |
| regulation of triglyceride biosynthetic process | 1 | -1.2 | 1.03 | 1.07 | -1.11 | 1.52 | -1.28 | -1.06 |
| lipid localization | 42 | -1.57 | -1.37 | -1.54 | 1.04 | -1.02 | -1.33 | -1.47 |
| regulation of lipid storage | 14 | 1.04 | 1.01 | -1.06 | 1.04 | 1.43 | -1.14 | 1.14 |
| positive regulation of lipid storage | 1 | -2.11 | 1.69 | -1.72 | -1.16 | 2.47 | -2.75 | -1.82 |
| negative regulation of lipid storage | 2 | 1.04 | 1.46 | 1.79 | 1.31 | 1.26 | 1.68 | 1.1 |
| regulation of sequestering of triglyceride | 2 | 1.65 | 1.42 | 1.72 | -1.12 | -1.11 | 1.9 | 1.54 |
| negative regulation of sequestering of triglyceride | 1 | 1.15 | 1.61 | -1.09 | 1.48 | -1.24 | 1.35 | -1.03 |
| regulation of glucose metabolic process | 2 | -1.67 | -1.02 | -1.57 | -1.22 | -1.25 | -1.34 | 1.83 |
| regulation of phosphatase activity | 3 | 1.09 | 1.05 | 1.0 | 1.06 | 1.03 | 1.05 | -1.09 |
| cellular component assembly | 455 | 1.19 | 1.06 | 1.24 | 1.05 | 1.23 | 1.24 | 1.12 |
| anatomical structure formation involved in morphogenesis | 388 | 1.14 | 1.06 | 1.21 | 1.03 | 1.22 | 1.22 | 1.17 |
| cellular component assembly involved in morphogenesis | 135 | 1.25 | 1.03 | 1.17 | 1.01 | 1.15 | 1.18 | 1.06 |
| regulation of cell death | 118 | 1.1 | 1.14 | 1.31 | 1.07 | 1.3 | 1.2 | 1.22 |
| positive regulation of cell death | 49 | -1.01 | 1.15 | 1.2 | 1.09 | 1.32 | 1.04 | 1.24 |
| negative regulation of cell cycle process | 32 | 1.06 | 1.12 | 1.28 | 1.15 | 1.41 | 1.18 | 1.22 |
| positive regulation of endopeptidase activity | 14 | 1.01 | 1.14 | 1.25 | 1.19 | 1.42 | 1.19 | 1.06 |
| negative regulation of endopeptidase activity | 3 | -1.25 | -1.08 | 1.67 | 1.12 | 1.52 | -1.68 | -1.23 |
| positive regulation of peptidase activity | 14 | 1.01 | 1.14 | 1.25 | 1.19 | 1.42 | 1.19 | 1.06 |
| negative regulation of protein processing | 1 | -2.9 | -1.14 | 1.64 | 1.56 | 2.18 | -4.01 | -1.31 |
| regulation of metal ion transport | 15 | 1.16 | 1.05 | 1.17 | 1.01 | 1.06 | 1.2 | 1.1 |
| magnesium ion homeostasis | 1 | -1.13 | 1.01 | 1.23 | 1.08 | 1.32 | 1.53 | 1.35 |
| regulation of microtubule nucleation | 1 | 2.49 | 1.3 | 2.87 | -1.0 | 2.26 | 3.13 | 2.77 |
| microtubule-based transport | 27 | -1.04 | 1.2 | 1.25 | 1.11 | 1.21 | 1.22 | 1.11 |
| positive regulation of G2/M transition of mitotic cell cycle | 1 | -1.03 | 1.05 | 1.22 | 1.04 | 1.19 | 1.13 | -1.03 |
| negative regulation of G2/M transition of mitotic cell cycle | 4 | -1.01 | -1.0 | 1.12 | 1.16 | 1.35 | 1.03 | -1.06 |
| regulation of neuron projection development | 47 | 1.05 | 1.15 | 1.4 | 1.08 | 1.39 | 1.28 | 1.2 |
| anaphase-promoting complex binding | 1 | 1.0 | -1.01 | -1.14 | -1.09 | -1.19 | -1.12 | -1.06 |
| regulation of translational initiation by eIF2 alpha phosphorylation | 3 | 3.45 | -1.5 | -1.27 | -1.43 | 1.18 | 2.31 | 1.49 |
| induction of programmed cell death | 35 | 1.04 | 1.13 | 1.16 | 1.09 | 1.31 | 1.02 | 1.25 |
| endomembrane system | 185 | 1.08 | 1.13 | 1.2 | 1.11 | 1.22 | 1.13 | 1.18 |
| ER to Golgi transport vesicle membrane | 1 | 1.02 | 1.56 | 2.15 | 1.06 | 2.07 | 2.36 | 2.5 |
| trans-Golgi network transport vesicle membrane | 3 | -1.23 | 1.16 | 1.44 | 1.05 | 1.28 | 1.6 | 1.31 |
| regulation of gliogenesis | 4 | 1.83 | 1.22 | 1.55 | -1.02 | 1.23 | 1.76 | 1.45 |
| negative regulation of gliogenesis | 2 | 1.15 | 1.57 | 1.36 | 1.13 | 1.31 | 1.77 | 1.58 |
| positive regulation of gliogenesis | 2 | 2.9 | -1.05 | 1.75 | -1.19 | 1.16 | 1.75 | 1.33 |
| neuroblast differentiation | 35 | 1.06 | 1.07 | 1.2 | 1.05 | 1.26 | 1.03 | 1.17 |
| neuroblast fate commitment | 27 | 1.15 | 1.09 | 1.19 | 1.07 | 1.25 | 1.03 | 1.13 |
| neuroblast fate specification | 1 | 2.48 | -1.28 | 1.14 | -1.16 | 1.81 | 2.29 | 3.17 |
| neuroblast development | 10 | -1.17 | 1.02 | 1.2 | 1.03 | 1.26 | 1.07 | 1.3 |
| mesenchymal cell development | 1 | 1.47 | 1.13 | -1.02 | 1.1 | -1.07 | -1.03 | 1.1 |
| glutamate secretion | 1 | -3.47 | -4.26 | 1.06 | 2.44 | 2.17 | -3.37 | -7.09 |
| postsynaptic density | 3 | 1.62 | -1.01 | 1.32 | -1.1 | 1.45 | 2.01 | 1.64 |
| response to organic cyclic compound | 31 | -1.0 | 1.1 | -1.1 | -1.05 | 1.23 | 1.17 | 1.37 |
| response to tropane | 13 | 1.28 | 1.17 | 1.57 | 1.07 | 1.39 | 1.83 | 1.27 |
| response to purine-containing compound | 4 | 1.2 | 1.25 | -1.32 | -1.41 | -1.17 | 1.21 | 1.42 |
| response to amine stimulus | 4 | 1.59 | -1.01 | 1.38 | 1.29 | 1.34 | 1.2 | -1.18 |
| oscillatory muscle contraction | 1 | -1.18 | -1.0 | 1.06 | -1.02 | -1.12 | 1.04 | 1.04 |
| striated muscle tissue development | 84 | 1.16 | 1.1 | 1.25 | 1.03 | 1.19 | 1.34 | 1.17 |
| muscle cell migration | 1 | 1.49 | 1.07 | 1.15 | 1.04 | -1.03 | 1.53 | -1.09 |
| regulation of skeletal muscle contraction | 1 | 2.24 | 1.81 | -38.59 | -31.15 | -18.58 | -3.21 | 4.55 |
| phasic smooth muscle contraction | 1 | 1.69 | 1.43 | 1.3 | 1.01 | 1.12 | 1.63 | -1.01 |
| regulation of skeletal muscle contraction by neural stimulation via neuromuscular junction | 1 | 2.24 | 1.81 | -38.59 | -31.15 | -18.58 | -3.21 | 4.55 |
| regulation of excitatory postsynaptic membrane potential involved in skeletal muscle contraction | 1 | 2.24 | 1.81 | -38.59 | -31.15 | -18.58 | -3.21 | 4.55 |
| skeletal myofibril assembly | 7 | 2.48 | -1.3 | 1.04 | -1.21 | 1.0 | 1.67 | -1.05 |
| myotube differentiation | 27 | 1.01 | 1.06 | 1.27 | -1.02 | 1.21 | 1.27 | 1.14 |
| heme-copper terminal oxidase activity | 18 | -1.12 | -1.08 | 1.07 | -1.07 | 1.03 | 1.19 | -1.31 |
| heparan sulfate proteoglycan biosynthetic process | 12 | 1.32 | 1.18 | 1.4 | 1.15 | 1.27 | 1.3 | 1.26 |
| heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 4 | 1.19 | 1.5 | 1.46 | 1.15 | 1.54 | 1.21 | 1.67 |
| [heparan sulfate]-glucosamine N-sulfotransferase activity | 1 | -1.1 | 1.79 | 1.14 | 1.04 | 1.02 | -1.23 | 2.55 |
| galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity | 3 | 1.19 | -1.06 | -1.32 | -1.33 | -1.21 | 1.07 | 3.7 |
| glucuronosyltransferase activity | 34 | -1.54 | -1.06 | -1.26 | 1.13 | 1.11 | -1.57 | -1.05 |
| anchored to membrane | 3 | 3.1 | -2.01 | -3.22 | -2.43 | -1.37 | 2.44 | 1.37 |
| Cajal body | 5 | 1.06 | 1.13 | 1.3 | 1.24 | 1.31 | 1.06 | -1.03 |
| protein transport | 243 | 1.06 | 1.16 | 1.3 | 1.12 | 1.23 | 1.13 | 1.15 |
| oxygen binding | 5 | 1.41 | 1.03 | 1.12 | -1.03 | -1.27 | 1.23 | -1.13 |
| protein disulfide oxidoreductase activity | 22 | 1.16 | 1.02 | 1.01 | 1.16 | 1.07 | -1.06 | -1.11 |
| peptide disulfide oxidoreductase activity | 6 | 1.07 | 1.19 | -1.04 | 1.07 | 1.14 | -1.12 | 1.05 |
| NADPH-adrenodoxin reductase activity | 1 | -1.19 | 1.24 | 1.2 | 1.43 | 1.53 | -1.12 | -1.13 |
| retinal binding | 6 | 3.18 | -5.84 | -5.89 | -2.35 | -1.77 | 1.79 | 1.62 |
| gastrin receptor activity | 2 | -1.2 | -1.05 | 1.02 | -1.05 | -1.23 | -1.25 | -1.17 |
| 1-phosphatidylinositol-3-kinase activity | 3 | -1.46 | 1.15 | 1.53 | 1.18 | 1.77 | -1.09 | -1.14 |
| 1-phosphatidylinositol-4-phosphate 3-kinase activity | 3 | -1.46 | 1.15 | 1.53 | 1.18 | 1.77 | -1.09 | -1.14 |
| phosphatidylinositol-4,5-bisphosphate 3-kinase activity | 3 | -1.46 | 1.15 | 1.53 | 1.18 | 1.77 | -1.09 | -1.14 |
| DNA integration | 1 | 1.07 | -1.04 | -1.16 | 1.02 | 1.48 | -1.27 | -1.22 |
| ion transmembrane transporter activity | 490 | -1.13 | -1.04 | -1.12 | -1.11 | -1.07 | 1.14 | -1.07 |
| monovalent inorganic cation transmembrane transporter activity | 190 | -1.23 | -1.03 | -1.22 | -1.16 | -1.14 | 1.14 | -1.18 |
| hydrogen ion transmembrane transporter activity | 82 | -1.19 | -1.01 | 1.17 | -1.07 | -1.05 | 1.38 | -1.22 |
| potassium ion transmembrane transporter activity | 49 | -1.01 | -1.08 | -1.08 | -1.16 | -1.05 | 1.13 | -1.06 |
| sodium ion transmembrane transporter activity | 81 | -1.48 | -1.05 | -1.89 | -1.26 | -1.27 | 1.0 | -1.16 |
| calcium ion transmembrane transporter activity | 11 | -1.32 | 1.18 | 1.08 | -1.02 | 1.12 | -1.09 | 1.13 |
| copper uptake transmembrane transporter activity | 1 | 1.39 | 1.37 | 1.47 | 1.19 | 1.52 | 2.16 | 1.06 |
| organic cation transmembrane transporter activity | 36 | -1.2 | -1.18 | -1.56 | -1.22 | -1.06 | 1.61 | -1.27 |
| inorganic anion transmembrane transporter activity | 44 | -1.75 | -1.05 | -1.79 | -1.14 | -1.41 | 1.18 | -1.54 |
| arsenite transmembrane transporter activity | 2 | 2.24 | 1.06 | 1.22 | -1.13 | -1.15 | 2.07 | 1.19 |
| bicarbonate transmembrane transporter activity | 1 | 1.2 | -1.03 | 1.07 | -1.04 | 1.13 | 1.1 | 1.23 |
| chloride transmembrane transporter activity | 4 | -2.12 | -1.32 | 1.67 | -1.4 | 1.05 | 1.49 | -1.53 |
| phosphate ion transmembrane transporter activity | 28 | -2.04 | -1.01 | -2.2 | -1.07 | -1.55 | -1.11 | -1.87 |
| sulfate transmembrane transporter activity | 11 | -1.38 | -1.18 | -1.62 | -1.28 | -1.18 | 2.41 | -1.09 |
| thiosulfate transmembrane transporter activity | 2 | 1.03 | -1.5 | 1.48 | -1.27 | 1.66 | 3.63 | -1.34 |
| bile acid transmembrane transporter activity | 2 | -3.13 | 1.41 | -2.05 | 1.29 | -3.5 | -4.18 | -1.28 |
| oxaloacetate transmembrane transporter activity | 1 | -1.02 | -2.78 | 1.86 | -1.91 | 2.75 | 13.01 | -1.81 |
| citrate transmembrane transporter activity | 6 | 1.66 | 1.03 | 1.2 | -1.17 | -1.14 | 1.49 | 1.13 |
| malate transmembrane transporter activity | 6 | -1.06 | -1.19 | 1.21 | -1.11 | 1.09 | 1.9 | -1.26 |
| succinate transmembrane transporter activity | 2 | 1.52 | -1.49 | 2.44 | -1.88 | 1.67 | 5.92 | 1.16 |
| urate transmembrane transporter activity | 2 | 1.45 | -1.07 | 1.88 | 1.03 | 1.06 | 1.31 | 1.14 |
| carbohydrate transmembrane transporter activity | 35 | -1.0 | -1.02 | -1.46 | -1.04 | -1.16 | -1.22 | 1.12 |
| monosaccharide transmembrane transporter activity | 21 | -1.07 | -1.11 | -1.82 | -1.2 | -1.34 | -1.42 | 1.07 |
| hexose transmembrane transporter activity | 18 | -1.2 | -1.08 | -1.91 | -1.2 | -1.46 | -1.81 | 1.09 |
| cocaine binding | 1 | -1.02 | 1.06 | -1.1 | 1.08 | -1.08 | 1.0 | -1.01 |
| disaccharide transmembrane transporter activity | 1 | 2.52 | -1.26 | -1.18 | -1.09 | -1.25 | 2.25 | 1.03 |
| pyrimidine nucleotide sugar transmembrane transporter activity | 8 | 1.01 | 1.23 | -1.01 | 1.24 | 1.15 | -1.17 | 1.39 |
| polyol transmembrane transporter activity | 1 | -2.04 | 1.18 | -1.02 | 1.21 | 1.32 | -1.45 | 1.81 |
| glycerol transmembrane transporter activity | 1 | -2.04 | 1.18 | -1.02 | 1.21 | 1.32 | -1.45 | 1.81 |
| glycerol-3-phosphate transmembrane transporter activity | 1 | -2.04 | 1.18 | -1.02 | 1.21 | 1.32 | -1.45 | 1.81 |
| acidic amino acid transmembrane transporter activity | 4 | -2.07 | 1.33 | -5.17 | -2.48 | -1.62 | -1.72 | -2.15 |
| aromatic amino acid transmembrane transporter activity | 2 | 3.0 | -1.1 | 1.15 | -1.28 | -1.42 | 2.49 | 1.13 |
| neutral amino acid transmembrane transporter activity | 5 | -1.05 | -1.63 | -1.94 | -1.23 | 1.36 | 1.17 | 1.17 |
| L-amino acid transmembrane transporter activity | 14 | -1.06 | -1.11 | -2.62 | -1.34 | 1.08 | -1.05 | -1.21 |
| L-aspartate transmembrane transporter activity | 2 | 1.6 | 1.38 | -6.52 | -5.62 | -4.41 | -1.75 | 2.45 |
| L-cystine transmembrane transporter activity | 1 | -7.18 | -1.45 | -9.47 | 1.62 | 3.49 | -2.41 | -5.84 |
| L-gamma-aminobutyric acid transmembrane transporter activity | 3 | 1.55 | 1.3 | -1.41 | -1.0 | 1.17 | -1.0 | 1.21 |
| glycine transmembrane transporter activity | 3 | -1.07 | 1.29 | 1.01 | -1.05 | -1.24 | -1.24 | 1.3 |
| L-proline transmembrane transporter activity | 1 | 1.01 | -1.09 | -1.18 | -1.18 | 1.11 | -1.08 | -1.12 |
| peptide transporter activity | 3 | 1.69 | -1.17 | -1.51 | -1.15 | -1.07 | 1.17 | 1.2 |
| oligopeptide transporter activity | 3 | 1.69 | -1.17 | -1.51 | -1.15 | -1.07 | 1.17 | 1.2 |
| nucleobase transmembrane transporter activity | 1 | 2.22 | 1.18 | 2.27 | 1.0 | 1.25 | 2.33 | 2.07 |
| purine nucleoside transmembrane transporter activity | 2 | 1.45 | 1.3 | 1.01 | 1.18 | 1.09 | 1.6 | 1.46 |
| choline transmembrane transporter activity | 1 | 1.01 | -1.09 | -1.18 | -1.18 | 1.11 | -1.08 | -1.12 |
| serotonin transmembrane transporter activity | 1 | -1.06 | -1.11 | -1.0 | -1.08 | -1.15 | 3.37 | -1.04 |
| vitamin transporter activity | 7 | -1.0 | 1.07 | -1.8 | -1.21 | 1.11 | -1.18 | 1.04 |
| cofactor transporter activity | 6 | -1.15 | 1.2 | 1.13 | 1.16 | 1.09 | -1.08 | 1.03 |
| carnitine transporter activity | 6 | -1.28 | 1.49 | -1.43 | -1.09 | -1.24 | -2.46 | -1.13 |
| acyl carnitine transporter activity | 2 | 1.23 | 1.21 | -1.11 | 1.11 | 1.06 | -1.22 | -1.1 |
| heme transporter activity | 4 | -1.09 | 1.3 | 1.25 | 1.23 | 1.07 | -1.15 | -1.06 |
| thiamine transmembrane transporter activity | 1 | 1.18 | 1.26 | -1.06 | 1.24 | -1.11 | -1.61 | 1.34 |
| drug transmembrane transporter activity | 14 | -1.4 | 1.1 | -1.22 | 1.07 | 1.18 | -1.23 | 1.11 |
| fatty acid transporter activity | 7 | -2.15 | 1.3 | -1.32 | -1.52 | -2.35 | -1.39 | -1.5 |
| sterol transporter activity | 1 | -1.06 | 1.09 | -1.0 | 1.4 | 1.63 | -1.48 | -1.12 |
| water channel activity | 5 | -2.72 | 1.35 | -1.04 | -1.24 | -3.15 | -2.7 | -1.32 |
| sugar transmembrane transporter activity | 26 | -1.05 | -1.08 | -1.66 | -1.14 | -1.28 | -1.32 | 1.13 |
| channel activity | 217 | 1.02 | 1.0 | -1.01 | -1.05 | -1.07 | 1.03 | -1.01 |
| calcium-activated potassium channel activity | 3 | -1.25 | -1.55 | 1.25 | 1.49 | -1.39 | -1.46 | -1.49 |
| outward rectifier potassium channel activity | 4 | 1.21 | 1.23 | 1.58 | -1.53 | -1.48 | 1.98 | -1.52 |
| ATP-activated inward rectifier potassium channel activity | 1 | -2.21 | 1.6 | 1.01 | 1.47 | -3.02 | -10.91 | -2.13 |
| ligand-gated ion channel activity | 108 | 1.02 | 1.0 | -1.0 | -1.08 | -1.14 | 1.0 | -1.02 |
| kainate selective glutamate receptor activity | 11 | 2.2 | -1.06 | 1.07 | -1.1 | -1.06 | 1.26 | 1.02 |
| calcium-release channel activity | 4 | 2.09 | 1.04 | 1.5 | 1.02 | 1.16 | 2.3 | 1.48 |
| store-operated calcium channel activity | 4 | 1.41 | -1.11 | 1.15 | 1.07 | 1.1 | 1.6 | 1.28 |
| ligand-gated sodium channel activity | 3 | 1.09 | -1.09 | -1.08 | -1.02 | -1.08 | -1.08 | -1.06 |
| porin activity | 1 | -1.24 | 1.02 | -1.02 | 1.01 | -1.17 | -1.1 | 1.01 |
| pore complex | 32 | 1.22 | 1.16 | 1.46 | 1.19 | 1.42 | 1.4 | 1.18 |
| symporter activity | 76 | -1.49 | -1.09 | -1.95 | -1.27 | -1.28 | -1.09 | -1.13 |
| solute:cation symporter activity | 74 | -1.49 | -1.1 | -1.98 | -1.28 | -1.29 | -1.08 | -1.14 |
| solute:hydrogen symporter activity | 10 | 1.17 | -1.04 | -1.27 | -1.11 | -1.11 | -1.04 | 1.72 |
| anion:cation symporter activity | 36 | -2.03 | -1.02 | -2.27 | -1.32 | -1.7 | -1.35 | -1.62 |
| antiporter activity | 25 | -1.4 | 1.02 | 1.06 | -1.09 | -1.06 | 1.2 | -1.02 |
| solute:cation antiporter activity | 13 | -1.59 | 1.16 | -1.1 | 1.0 | -1.16 | 1.34 | 1.01 |
| solute:hydrogen antiporter activity | 6 | -1.84 | 1.01 | -1.18 | -1.0 | -1.46 | 2.19 | -1.2 |
| solute:solute antiporter activity | 24 | -1.43 | 1.01 | 1.06 | -1.1 | -1.08 | 1.21 | -1.06 |
| anion:anion antiporter activity | 7 | -1.5 | -1.45 | 1.6 | -1.5 | -1.11 | 1.16 | -1.09 |
| organophosphate:inorganic phosphate antiporter activity | 1 | 1.19 | 1.35 | 1.39 | 1.1 | 1.22 | 1.13 | 2.76 |
| sodium:inorganic phosphate symporter activity | 1 | -51.3 | 1.56 | -15.93 | -1.24 | 2.78 | -3.27 | -134.18 |
| sodium-dependent phosphate transmembrane transporter activity | 1 | -1.14 | 1.17 | 1.24 | 1.15 | -1.13 | -1.05 | 2.26 |
| secondary active oligopeptide transmembrane transporter activity | 3 | 1.69 | -1.17 | -1.51 | -1.15 | -1.07 | 1.17 | 1.2 |
| cationic amino acid transmembrane transporter activity | 5 | -2.27 | 1.25 | -1.58 | -1.02 | 1.06 | 1.04 | -1.06 |
| sodium-independent organic anion transmembrane transporter activity | 3 | -1.97 | -1.09 | 1.04 | 1.04 | 1.56 | -1.02 | -1.01 |
| secondary active monocarboxylate transmembrane transporter activity | 6 | -2.96 | -1.49 | -1.43 | 1.36 | 1.66 | 1.29 | -1.83 |
| calcium:cation antiporter activity | 7 | -1.41 | 1.31 | -1.04 | 1.01 | 1.05 | -1.14 | 1.2 |
| solute:sodium symporter activity | 58 | -1.57 | -1.09 | -2.44 | -1.32 | -1.33 | -1.17 | -1.25 |
| monovalent anion:sodium symporter activity | 7 | 1.13 | -1.12 | -1.82 | -1.93 | -1.54 | -1.43 | 1.18 |
| glycine:sodium symporter activity | 1 | -1.01 | 1.0 | 1.27 | 1.1 | -1.12 | 1.05 | -1.06 |
| cation:chloride symporter activity | 4 | -2.12 | -1.32 | 1.67 | -1.4 | 1.05 | 1.49 | -1.53 |
| potassium:chloride symporter activity | 1 | -4.23 | -1.01 | 5.33 | -1.14 | 1.37 | -4.42 | -4.03 |
| membrane docking | 27 | -1.1 | 1.16 | 1.4 | 1.15 | 1.26 | 1.14 | 1.1 |
| high affinity sulfate transmembrane transporter activity | 8 | -1.57 | -1.12 | -2.1 | -1.3 | -1.39 | 2.29 | -1.04 |
| sodium:hydrogen antiporter activity | 4 | -2.37 | -1.01 | -1.35 | 1.01 | -1.74 | 3.33 | -1.27 |
| nucleoside transmembrane transporter activity, down a concentration gradient | 1 | 1.01 | 1.49 | 1.07 | 1.13 | 1.13 | 1.28 | 1.42 |
| primary active transmembrane transporter activity | 143 | -1.07 | -1.01 | 1.05 | -1.04 | 1.03 | 1.15 | -1.05 |
| P-P-bond-hydrolysis-driven transmembrane transporter activity | 143 | -1.07 | -1.01 | 1.05 | -1.04 | 1.03 | 1.15 | -1.05 |
| manganese-transporting ATPase activity | 1 | 1.04 | 1.04 | -1.04 | -1.09 | -1.16 | -1.14 | -1.1 |
| arsenite-transmembrane transporting ATPase activity | 1 | 1.35 | 1.29 | 1.36 | 1.08 | 1.19 | 1.34 | 1.11 |
| P-P-bond-hydrolysis-driven protein transmembrane transporter activity | 23 | -1.01 | 1.13 | 1.18 | 1.11 | 1.07 | 1.12 | -1.06 |
| potassium channel regulator activity | 2 | 1.41 | 1.19 | 1.44 | 1.03 | 1.46 | 1.62 | 1.3 |
| acetylcholine receptor activity | 2 | 1.0 | 1.08 | 1.1 | 1.16 | 1.17 | 1.06 | 1.05 |
| G-protein activated inward rectifier potassium channel activity | 1 | 4.33 | -4.87 | -19.75 | -19.08 | -6.11 | 18.23 | -1.15 |
| cholesterol binding | 2 | 1.08 | 1.11 | 1.07 | 1.26 | 1.29 | -1.19 | -1.05 |
| cation:cation antiporter activity | 11 | -1.7 | 1.18 | -1.14 | 1.01 | -1.19 | 1.42 | 1.03 |
| gamma-aminobutyric acid:hydrogen symporter activity | 2 | 1.74 | 1.43 | -1.72 | -1.09 | 1.17 | -1.0 | 1.38 |
| glutamate:sodium symporter activity | 2 | 1.6 | 1.38 | -6.52 | -5.62 | -4.41 | -1.75 | 2.45 |
| C4-dicarboxylate transmembrane transporter activity | 7 | 1.08 | -1.13 | 1.39 | -1.2 | 1.08 | 1.99 | -1.07 |
| thiamine pyrophosphate-transporting ATPase activity | 1 | 1.18 | 1.26 | -1.06 | 1.24 | -1.11 | -1.61 | 1.34 |
| actin cytoskeleton | 94 | -1.06 | -1.02 | 1.2 | 1.01 | 1.23 | 1.08 | -1.0 |
| microtubule cytoskeleton | 439 | 1.11 | 1.04 | 1.13 | 1.04 | 1.19 | 1.16 | 1.06 |
| tubulin binding | 67 | 1.1 | 1.03 | 1.32 | 1.0 | 1.34 | 1.37 | 1.25 |
| fatty acid ligase activity | 7 | 1.81 | -1.3 | -1.35 | 1.09 | 1.2 | -1.91 | -1.26 |
| quaternary ammonium group transmembrane transporter activity | 6 | -1.28 | 1.49 | -1.43 | -1.09 | -1.24 | -2.46 | -1.13 |
| alcohol transmembrane transporter activity | 2 | -1.44 | 1.12 | -1.06 | 1.14 | 1.1 | -1.2 | 1.33 |
| gas transport | 3 | 2.18 | 1.01 | 1.23 | -1.11 | -1.43 | 1.5 | -1.22 |
| oxygen transport | 3 | 2.18 | 1.01 | 1.23 | -1.11 | -1.43 | 1.5 | -1.22 |
| monovalent inorganic cation transport | 136 | -1.06 | -1.02 | 1.06 | -1.08 | -1.06 | 1.22 | -1.09 |
| copper ion import | 2 | -1.71 | 1.19 | -1.03 | 1.03 | -1.3 | -1.51 | 1.21 |
| plasma membrane copper ion transport | 1 | 1.39 | 1.37 | 1.47 | 1.19 | 1.52 | 2.16 | 1.06 |
| intracellular copper ion transport | 1 | 1.35 | 1.23 | -1.1 | 1.09 | -1.09 | -1.29 | 1.14 |
| cadmium ion transport | 1 | -1.37 | 1.35 | 1.68 | 1.28 | 1.12 | -1.35 | -1.15 |
| organic cation transport | 6 | -1.21 | -1.29 | -1.94 | -1.42 | -1.18 | 1.53 | 1.11 |
| ammonium transport | 1 | -1.13 | -1.02 | -1.13 | -1.06 | -1.15 | -1.01 | -1.16 |
| quaternary ammonium group transport | 2 | 1.34 | 1.32 | -1.91 | 1.22 | 1.14 | -1.7 | -1.23 |
| inorganic anion transport | 18 | -1.11 | 1.04 | -1.32 | -1.05 | 1.11 | 1.94 | -1.22 |
| bicarbonate transport | 1 | 1.2 | -1.03 | 1.07 | -1.04 | 1.13 | 1.1 | 1.23 |
| thiosulfate transport | 2 | 1.03 | -1.5 | 1.48 | -1.27 | 1.66 | 3.63 | -1.34 |
| organic anion transport | 23 | -1.44 | -1.02 | -1.33 | -1.21 | -1.04 | -1.01 | -1.2 |
| monocarboxylic acid transport | 2 | 1.54 | 1.0 | 1.06 | 1.2 | -1.29 | 1.7 | 1.68 |
| oxaloacetate transport | 1 | -1.02 | -2.78 | 1.86 | -1.91 | 2.75 | 13.01 | -1.81 |
| taurine transport | 2 | -1.75 | -2.01 | -1.02 | 1.55 | 1.44 | -1.8 | -2.32 |
| C4-dicarboxylate transport | 5 | -1.06 | -1.22 | 1.26 | -1.12 | 1.12 | 2.23 | -1.26 |
| succinate transport | 1 | -1.02 | -2.78 | 1.86 | -1.91 | 2.75 | 13.01 | -1.81 |
| citrate transport | 6 | 1.38 | 1.1 | -1.07 | -1.11 | -1.31 | 1.27 | -1.04 |
| monosaccharide transport | 5 | 1.14 | -1.11 | -1.24 | -1.1 | 1.05 | 1.47 | 1.25 |
| galactose transport | 1 | -3.25 | 1.38 | -1.03 | -1.03 | -1.21 | -3.18 | 3.55 |
| glucose transport | 1 | -1.11 | -1.03 | -1.12 | 1.1 | -1.16 | -1.22 | -1.12 |
| disaccharide transport | 3 | 1.57 | -1.8 | -1.19 | -1.63 | -1.17 | 2.95 | 1.2 |
| sucrose transport | 1 | 2.52 | -1.26 | -1.18 | -1.09 | -1.25 | 2.25 | 1.03 |
| trehalose transport | 2 | 1.24 | -2.16 | -1.19 | -2.0 | -1.13 | 3.38 | 1.3 |
| oligosaccharide transport | 3 | 1.57 | -1.8 | -1.19 | -1.63 | -1.17 | 2.95 | 1.2 |
| nucleotide-sugar transport | 4 | -1.46 | 1.25 | 1.0 | 1.26 | 1.04 | -1.57 | 1.43 |
| pyrimidine nucleotide-sugar transport | 4 | -1.46 | 1.25 | 1.0 | 1.26 | 1.04 | -1.57 | 1.43 |
| CMP-N-acetylneuraminate transport | 3 | -1.73 | 1.25 | -1.03 | 1.28 | -1.06 | -1.84 | 1.23 |
| GDP-fucose transport | 2 | -1.26 | 1.19 | -1.04 | 1.47 | 1.01 | -1.41 | -1.39 |
| GDP-mannose transport | 2 | -1.26 | 1.19 | -1.04 | 1.47 | 1.01 | -1.41 | -1.39 |
| UDP-galactose transport | 3 | -1.67 | 1.25 | -1.02 | 1.2 | 1.18 | -1.82 | 1.71 |
| UDP-glucose transport | 3 | -1.11 | 1.21 | 1.01 | 1.37 | 1.12 | -1.24 | 1.06 |
| UDP-glucuronic acid transport | 3 | -1.11 | 1.21 | 1.01 | 1.37 | 1.12 | -1.24 | 1.06 |
| UDP-N-acetylglucosamine transport | 3 | -1.11 | 1.21 | 1.01 | 1.37 | 1.12 | -1.24 | 1.06 |
| UDP-N-acetylgalactosamine transport | 2 | -2.31 | 1.25 | -1.08 | 1.21 | 1.09 | -2.5 | 1.48 |
| UDP-xylose transport | 2 | 1.09 | 1.26 | 1.09 | 1.3 | 1.0 | 1.02 | 1.39 |
| polyol transport | 1 | -1.32 | 1.18 | 1.15 | 1.08 | 1.65 | -1.36 | 1.01 |
| myo-inositol transport | 1 | -1.32 | 1.18 | 1.15 | 1.08 | 1.65 | -1.36 | 1.01 |
| acidic amino acid transport | 4 | -2.92 | -1.1 | -4.98 | -1.98 | -1.32 | -2.35 | -3.75 |
| branched-chain aliphatic amino acid transport | 3 | 1.2 | -1.03 | -1.17 | 1.06 | 1.15 | -1.13 | 2.49 |
| neutral amino acid transport | 6 | -1.07 | -2.12 | -1.83 | 1.1 | 1.71 | -1.03 | 1.19 |
| L-amino acid transport | 3 | -7.27 | -2.77 | -15.38 | -1.15 | 4.25 | -1.2 | -8.88 |
| L-cystine transport | 1 | -7.18 | -1.45 | -9.47 | 1.62 | 3.49 | -2.41 | -5.84 |
| leucine transport | 3 | 1.2 | -1.03 | -1.17 | 1.06 | 1.15 | -1.13 | 2.49 |
| ornithine transport | 1 | 1.32 | 1.45 | -1.5 | 1.71 | 1.05 | 1.03 | -1.1 |
| proline transport | 1 | 1.41 | 1.17 | -1.02 | 1.18 | -1.05 | -1.09 | 1.18 |
| peptide transport | 4 | 1.61 | -1.15 | -1.24 | -1.1 | 1.08 | 1.07 | 1.44 |
| amine transport | 43 | -1.38 | -1.14 | -1.77 | -1.23 | -1.02 | -1.05 | -1.16 |
| synaptic vesicle amine transport | 1 | 9.63 | -2.21 | -1.97 | -1.79 | -1.94 | -1.46 | -1.99 |
| monoamine transport | 3 | 2.07 | -1.32 | -1.3 | -1.21 | -1.34 | 1.32 | -1.28 |
| organic acid transport | 54 | -1.28 | -1.1 | -1.53 | -1.18 | -1.03 | 1.07 | -1.12 |
| organic alcohol transport | 3 | 1.93 | -1.21 | -1.24 | -1.15 | -1.08 | -1.25 | -1.26 |
| nucleoside transport | 3 | 1.64 | 1.2 | 1.17 | 1.2 | 1.5 | 1.69 | 1.3 |
| purine nucleoside transport | 1 | 1.9 | 1.27 | 1.23 | 1.06 | 1.26 | 2.28 | 1.58 |
| purine nucleotide transport | 2 | -1.19 | 1.19 | 1.41 | 1.1 | 1.28 | 1.45 | -1.24 |
| ADP transport | 2 | -1.19 | 1.19 | 1.41 | 1.1 | 1.28 | 1.45 | -1.24 |
| ATP transport | 2 | -1.19 | 1.19 | 1.41 | 1.1 | 1.28 | 1.45 | -1.24 |
| purine ribonucleotide transport | 2 | -1.19 | 1.19 | 1.41 | 1.1 | 1.28 | 1.45 | -1.24 |
| dopamine transport | 2 | 3.07 | -1.44 | -1.47 | -1.28 | -1.45 | -1.2 | -1.42 |
| vitamin transport | 5 | -1.7 | 1.08 | -1.68 | 1.05 | 1.16 | -1.37 | -1.11 |
| cofactor transport | 6 | -1.04 | 1.23 | -1.1 | 1.14 | 1.09 | -1.22 | -1.05 |
| carnitine transport | 2 | 1.34 | 1.32 | -1.91 | 1.22 | 1.14 | -1.7 | -1.23 |
| folic acid transport | 1 | -1.3 | -1.21 | 1.08 | -1.14 | 1.42 | 1.82 | -1.08 |
| heme transport | 2 | -1.43 | 1.37 | 1.44 | 1.16 | 1.01 | -1.14 | -1.05 |
| thiamine transport | 1 | 1.18 | 1.26 | -1.06 | 1.24 | -1.11 | -1.61 | 1.34 |
| cobalamin transport | 1 | -1.04 | 1.01 | 1.04 | 1.07 | -1.1 | -1.21 | 1.08 |
| drug transport | 5 | -1.71 | 1.01 | -2.04 | -1.18 | 1.46 | -1.6 | -1.08 |
| fatty acid transport | 2 | 1.54 | 1.0 | 1.06 | 1.2 | -1.29 | 1.7 | 1.68 |
| long-chain fatty acid transport | 2 | 1.54 | 1.0 | 1.06 | 1.2 | -1.29 | 1.7 | 1.68 |
| phospholipid transport | 7 | -2.51 | 1.11 | -1.27 | 1.27 | 1.28 | -1.64 | -1.28 |
| sterol transport | 9 | -4.78 | -2.96 | -3.21 | 1.02 | -1.76 | -3.24 | -5.0 |
| aspartate oxidase activity | 2 | -2.82 | -1.69 | -2.22 | 1.7 | 1.95 | -1.63 | -1.83 |
| mannosidase activity | 15 | -4.86 | 1.01 | -3.54 | 1.28 | -1.09 | -4.24 | -6.43 |
| mannosyl-oligosaccharide mannosidase activity | 7 | 1.72 | 1.11 | -1.23 | -1.05 | -1.05 | 2.48 | 1.36 |
| galactosidase activity | 3 | -4.2 | 1.61 | -2.03 | -1.24 | -2.09 | -1.97 | -1.23 |
| glucosidase activity | 16 | -6.01 | -1.16 | -3.27 | 1.15 | -2.07 | -4.27 | -2.24 |
| trehalase activity | 3 | -2.73 | 1.23 | -2.68 | 1.17 | -1.22 | -2.41 | 1.01 |
| fucosidase activity | 1 | -1.1 | -1.19 | -1.53 | -1.9 | -1.86 | -1.28 | 7.99 |
| hexosaminidase activity | 7 | 1.77 | -1.0 | -1.28 | -1.06 | 1.05 | 1.54 | 1.4 |
| glutamate synthase activity | 2 | 2.76 | -1.53 | 2.1 | -1.11 | -1.64 | -1.33 | 1.55 |
| nucleobase-containing compound transport | 53 | -1.03 | 1.16 | 1.23 | 1.13 | 1.28 | 1.19 | 1.02 |
| nucleobase-containing compound transmembrane transporter activity | 7 | -1.27 | 1.29 | -1.17 | 1.12 | -1.04 | -1.15 | 1.06 |
| large ribosomal subunit | 95 | 1.2 | 1.04 | 1.16 | 1.13 | 1.2 | 1.31 | -1.19 |
| small ribosomal subunit | 62 | 1.2 | 1.05 | 1.17 | 1.13 | 1.19 | 1.3 | -1.18 |
| coenzyme A metabolic process | 5 | 1.35 | 1.24 | 1.03 | 1.25 | 1.35 | -1.15 | -1.04 |
| coenzyme A biosynthetic process | 5 | 1.35 | 1.24 | 1.03 | 1.25 | 1.35 | -1.15 | -1.04 |
| methane metabolic process | 1 | 1.08 | 1.05 | 1.26 | 1.07 | 1.31 | 1.33 | 1.1 |
| methanogenesis | 1 | 1.08 | 1.05 | 1.26 | 1.07 | 1.31 | 1.33 | 1.1 |
| nucleobase-containing small molecule interconversion | 1 | 2.25 | -1.1 | -1.03 | 1.28 | 1.09 | 2.14 | -1.24 |
| pyrimidine nucleotide interconversion | 1 | 2.25 | -1.1 | -1.03 | 1.28 | 1.09 | 2.14 | -1.24 |
| pyrimidine deoxyribonucleotide interconversion | 1 | 2.25 | -1.1 | -1.03 | 1.28 | 1.09 | 2.14 | -1.24 |
| guanosine tetraphosphate metabolic process | 1 | 1.81 | -1.03 | 1.57 | 1.17 | 1.71 | 1.48 | -1.14 |
| guanosine tetraphosphate catabolic process | 1 | 1.81 | -1.03 | 1.57 | 1.17 | 1.71 | 1.48 | -1.14 |
| energy derivation by oxidation of reduced inorganic compounds | 1 | 1.08 | 1.05 | 1.26 | 1.07 | 1.31 | 1.33 | 1.1 |
| carbon utilization | 1 | 3.68 | 1.14 | -1.19 | 1.01 | 1.66 | 1.38 | 1.25 |
| energy derivation by oxidation of organic compounds | 106 | 1.09 | 1.04 | 1.19 | 1.01 | 1.12 | 1.62 | -1.17 |
| proton transport | 51 | -1.18 | 1.0 | 1.25 | -1.1 | -1.04 | 1.38 | -1.15 |
| energy coupled proton transport, down electrochemical gradient | 18 | 1.37 | -1.0 | 1.2 | -1.02 | 1.12 | 1.7 | -1.2 |
| ATP synthesis coupled proton transport | 18 | 1.37 | -1.0 | 1.2 | -1.02 | 1.12 | 1.7 | -1.2 |
| energy coupled proton transport, against electrochemical gradient | 32 | -1.37 | -1.01 | 1.29 | -1.18 | -1.14 | 1.31 | -1.13 |
| ATP hydrolysis coupled proton transport | 32 | -1.37 | -1.01 | 1.29 | -1.18 | -1.14 | 1.31 | -1.13 |
| phospholipase activator activity | 2 | -1.41 | 1.32 | 1.65 | 1.18 | 1.23 | 1.2 | -1.03 |
| phospholipase A2 activator activity | 1 | -1.23 | 1.24 | 1.06 | 1.3 | 1.12 | 1.05 | -1.17 |
| Nebenkern | 6 | -1.27 | 1.23 | 1.24 | 1.01 | -1.05 | 1.16 | -1.05 |
| mitochondrial derivative | 3 | 1.07 | 1.03 | -1.0 | 1.03 | -1.06 | -1.03 | -1.02 |
| dystrophin-associated glycoprotein complex | 8 | 1.62 | 1.02 | 1.14 | 1.04 | 1.13 | 1.2 | 1.02 |
| dystroglycan complex | 4 | 2.12 | 1.06 | 1.24 | 1.03 | 1.2 | 1.2 | 1.03 |
| sarcoglycan complex | 3 | 2.49 | -1.02 | 1.02 | 1.01 | 1.11 | 1.14 | -1.09 |
| syntrophin complex | 2 | -1.29 | -1.06 | 1.25 | 1.01 | 1.12 | -1.18 | 1.08 |
| dystrobrevin complex | 1 | 4.45 | -1.13 | -1.95 | -1.08 | 1.28 | 3.01 | -1.11 |
| morphogen activity | 7 | 2.09 | -1.17 | 1.11 | -1.12 | 1.1 | 1.31 | 1.31 |
| peptidoglycan receptor activity | 3 | 2.74 | -1.01 | 1.12 | -1.1 | 1.04 | 2.17 | 1.15 |
| cytoplasmic membrane-bounded vesicle | 103 | 1.04 | 1.04 | 1.12 | 1.04 | 1.15 | 1.13 | 1.16 |
| CDP-diacylglycerol biosynthetic process | 1 | 3.07 | 1.1 | 2.07 | -1.42 | 3.13 | -1.11 | 1.4 |
| inaD signaling complex | 9 | -1.03 | -1.05 | 1.17 | 1.01 | -1.08 | -1.04 | -1.07 |
| rhabdomere | 27 | -1.04 | 1.03 | 1.2 | 1.01 | 1.03 | -1.05 | 1.0 |
| subrhabdomeral cisterna | 3 | 2.2 | -1.44 | 1.68 | 1.01 | -1.15 | 1.14 | 1.11 |
| maleylacetoacetate isomerase activity | 2 | 1.58 | 1.0 | 1.35 | 1.16 | 1.16 | 1.96 | -1.03 |
| zeta DNA polymerase complex | 1 | -1.03 | 1.1 | 1.04 | -1.05 | 1.06 | 2.37 | 1.1 |
| light absorption | 2 | -1.17 | 1.01 | -1.19 | -1.1 | -1.37 | -1.21 | -1.18 |
| absorption of visible light | 1 | -1.32 | 1.03 | -1.29 | -1.1 | -1.47 | -1.43 | -1.25 |
| absorption of UV light | 1 | -1.04 | -1.01 | -1.1 | -1.1 | -1.27 | -1.03 | -1.11 |
| glutamate synthase (NADH) activity | 1 | 2.14 | 1.19 | 1.24 | 1.03 | -1.73 | 3.68 | 2.51 |
| lipid catabolic process | 41 | -1.2 | -1.2 | -1.43 | 1.05 | 1.03 | -1.9 | -1.34 |
| cellular component organization | 1674 | 1.17 | 1.07 | 1.21 | 1.06 | 1.23 | 1.22 | 1.13 |
| cellular membrane organization | 260 | 1.07 | 1.1 | 1.21 | 1.08 | 1.23 | 1.18 | 1.13 |
| detection of bacterium | 5 | -1.12 | -2.27 | 1.01 | 1.08 | -1.9 | -1.65 | -3.19 |
| detection of temperature stimulus | 2 | 1.0 | 1.06 | -1.05 | 1.0 | -1.03 | -1.07 | -1.17 |
| vesicle organization | 37 | -1.12 | 1.13 | 1.35 | 1.17 | 1.24 | 1.09 | 1.12 |
| carbohydrate biosynthetic process | 54 | 1.37 | -1.06 | -1.04 | -1.09 | 1.09 | 1.31 | 1.23 |
| carbohydrate catabolic process | 70 | 1.14 | -1.3 | -1.52 | -1.28 | -1.28 | 1.14 | 1.27 |
| organic acid biosynthetic process | 65 | 1.05 | -1.1 | -1.35 | -1.11 | -1.17 | 1.1 | 1.09 |
| organic acid catabolic process | 40 | 1.0 | -1.07 | -1.2 | -1.06 | 1.05 | 1.14 | -1.23 |
| rhodopsin mediated signaling pathway | 9 | 1.31 | -1.0 | 1.3 | -1.05 | 1.29 | 1.08 | 1.04 |
| regulation of membrane potential in photoreceptor cell | 4 | -1.04 | 1.04 | 1.07 | -1.03 | 1.02 | 1.09 | -1.02 |
| maintenance of membrane potential in photoreceptor cell by rhodopsin mediated signaling | 1 | -1.2 | -1.13 | -1.12 | -1.01 | -1.14 | -1.07 | -1.41 |
| deactivation of rhodopsin mediated signaling | 13 | 1.3 | 1.02 | 1.13 | 1.05 | 1.11 | 1.08 | 1.05 |
| metarhodopsin inactivation | 4 | 1.14 | 1.05 | 1.18 | 1.1 | 1.25 | 1.13 | 1.01 |
| regulation of light-activated channel activity | 1 | -1.83 | 1.06 | 1.22 | 1.16 | 1.68 | -1.18 | -1.24 |
| adaptation of rhodopsin mediated signaling | 4 | -1.08 | 1.14 | 1.07 | 1.08 | 1.11 | -1.0 | -1.04 |
| rhodopsin biosynthetic process | 8 | -1.84 | -1.59 | -2.1 | 1.54 | 1.04 | -1.85 | -2.07 |
| immunoglobulin mediated immune response | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| type I hypersensitivity | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| RNA metabolic process | 549 | 1.22 | 1.14 | 1.23 | 1.19 | 1.27 | 1.18 | 1.08 |
| mRNA metabolic process | 247 | 1.19 | 1.12 | 1.23 | 1.21 | 1.31 | 1.2 | 1.04 |
| rRNA metabolic process | 29 | 1.2 | 1.17 | 1.21 | 1.31 | 1.31 | 1.05 | 1.06 |
| snRNA metabolic process | 2 | -1.23 | 1.23 | 1.84 | 1.26 | 1.56 | -1.27 | 1.05 |
| rRNA catabolic process | 1 | 1.49 | 1.15 | 1.39 | 1.19 | 1.14 | 2.46 | 1.32 |
| synaptic vesicle exocytosis | 49 | -1.1 | 1.08 | 1.28 | 1.15 | 1.24 | 1.11 | 1.09 |
| synaptic vesicle docking involved in exocytosis | 20 | -1.08 | 1.18 | 1.42 | 1.17 | 1.28 | 1.13 | 1.1 |
| synaptic vesicle priming | 13 | -1.03 | 1.07 | 1.48 | 1.09 | 1.32 | 1.19 | 1.13 |
| myostimulatory hormone activity | 1 | -1.18 | 1.06 | 1.01 | 1.05 | -1.08 | -1.09 | -1.08 |
| myoinhibitory hormone activity | 2 | -14.64 | -2.67 | -1.05 | 2.16 | -1.19 | -12.67 | -5.89 |
| negative regulation of juvenile hormone biosynthetic process | 1 | -84.21 | -3.59 | 1.42 | 2.07 | 1.14 | -58.36 | -15.07 |
| ecdysiostatic hormone activity | 1 | -2.55 | -1.98 | -1.56 | 2.24 | -1.62 | -2.75 | -2.3 |
| positive regulation of translation | 11 | 1.29 | 1.21 | 1.33 | 1.03 | 1.33 | 1.35 | 1.45 |
| positive regulation of nucleobase-containing compound metabolic process | 222 | 1.11 | 1.05 | 1.13 | 1.03 | 1.13 | 1.18 | 1.15 |
| positive regulation of lipid biosynthetic process | 1 | -3.9 | 1.39 | 1.21 | 1.36 | 1.18 | -2.17 | -1.43 |
| prenol metabolic process | 1 | 4.21 | 1.14 | 2.47 | -1.3 | 1.39 | 2.06 | 1.87 |
| prenol biosynthetic process | 1 | 4.21 | 1.14 | 2.47 | -1.3 | 1.39 | 2.06 | 1.87 |
| diterpenoid metabolic process | 3 | 1.01 | -1.01 | 1.02 | 1.05 | -1.08 | 1.07 | -1.1 |
| sesquiterpenoid biosynthetic process | 2 | 1.77 | -2.92 | -3.1 | -3.09 | -1.69 | -1.43 | 2.62 |
| sesquiterpenoid catabolic process | 4 | -1.67 | 1.21 | -1.1 | -1.05 | -1.31 | -3.02 | 1.26 |
| tetraterpenoid metabolic process | 3 | -1.97 | -1.44 | -2.29 | 1.38 | 1.12 | -1.8 | -2.35 |
| tetraterpenoid biosynthetic process | 1 | -1.0 | 1.21 | -1.05 | 1.12 | 1.06 | -1.02 | -1.03 |
| terpenoid biosynthetic process | 3 | 1.46 | -1.92 | -2.16 | -2.04 | -1.39 | -1.28 | 1.88 |
| terpenoid catabolic process | 4 | -1.67 | 1.21 | -1.1 | -1.05 | -1.31 | -3.02 | 1.26 |
| carotenoid metabolic process | 3 | -1.97 | -1.44 | -2.29 | 1.38 | 1.12 | -1.8 | -2.35 |
| carotenoid biosynthetic process | 1 | -1.0 | 1.21 | -1.05 | 1.12 | 1.06 | -1.02 | -1.03 |
| carotene metabolic process | 1 | 1.12 | -1.01 | 1.14 | 1.06 | 1.09 | 1.14 | -1.06 |
| sterol metabolic process | 14 | 1.26 | 1.04 | 1.06 | 1.05 | 1.15 | 1.45 | 1.17 |
| sterol biosynthetic process | 8 | 1.11 | 1.02 | 1.04 | 1.06 | -1.03 | 1.15 | 1.03 |
| sterol catabolic process | 1 | -1.19 | 1.24 | 1.2 | 1.43 | 1.53 | -1.12 | -1.13 |
| glycoside metabolic process | 7 | 1.73 | -1.53 | -1.79 | -1.54 | -1.23 | 1.97 | 1.32 |
| glycoside biosynthetic process | 3 | 3.57 | -2.83 | -2.37 | -2.36 | -1.16 | 5.45 | -1.46 |
| glycoside catabolic process | 2 | -1.06 | -1.02 | -1.16 | -1.17 | -1.27 | -1.11 | 2.75 |
| O-glycoside metabolic process | 2 | -1.06 | -1.02 | -1.16 | -1.17 | -1.27 | -1.11 | 2.75 |
| O-glycoside catabolic process | 2 | -1.06 | -1.02 | -1.16 | -1.17 | -1.27 | -1.11 | 2.75 |
| translation release factor activity, codon specific | 2 | 1.29 | 1.19 | 1.11 | 1.18 | 1.06 | 1.64 | 1.05 |
| formyltetrahydrofolate dehydrogenase activity | 1 | -1.35 | 1.22 | 1.7 | 1.18 | 1.15 | -1.09 | 1.03 |
| amylase activity | 2 | -34.84 | -2.22 | -3.89 | 2.13 | -10.58 | -52.06 | -2.46 |
| superoxide-generating NADPH oxidase activity | 1 | 2.71 | -1.01 | 1.01 | -1.06 | -1.03 | 2.12 | 1.84 |
| synaptic vesicle coating | 12 | 1.12 | 1.15 | 1.39 | 1.12 | 1.27 | 1.41 | 1.31 |
| synaptic vesicle budding from presynaptic membrane | 14 | 1.04 | 1.17 | 1.47 | 1.12 | 1.28 | 1.36 | 1.24 |
| synaptic vesicle maturation | 1 | 1.21 | 1.12 | 1.66 | 1.2 | 1.79 | 2.51 | 1.99 |
| endosome transport | 27 | 1.15 | 1.15 | 1.49 | 1.14 | 1.32 | 1.29 | 1.14 |
| axon choice point recognition | 24 | 1.17 | 1.11 | 1.3 | 1.03 | 1.22 | 1.12 | 1.17 |
| axon midline choice point recognition | 19 | 1.15 | 1.12 | 1.41 | 1.02 | 1.3 | 1.17 | 1.22 |
| synaptic target attraction | 3 | 1.93 | -1.35 | 1.08 | -1.19 | 1.21 | 2.92 | 1.18 |
| synaptic target inhibition | 1 | -1.51 | -1.03 | 1.25 | 1.21 | 2.96 | -2.96 | 1.02 |
| regulation of striated muscle tissue development | 47 | 1.01 | 1.1 | 1.22 | 1.05 | 1.15 | 1.1 | 1.16 |
| determination of muscle attachment site | 4 | 1.72 | 1.1 | 1.18 | 1.13 | 1.38 | 1.37 | 1.04 |
| selenocysteine methyltransferase activity | 2 | -2.66 | 1.46 | -3.05 | 1.35 | 1.97 | -1.55 | -1.14 |
| 4-coumarate-CoA ligase activity | 5 | -15.09 | -1.26 | -3.1 | -1.62 | -4.07 | -3.87 | -5.45 |
| AMP binding | 1 | 3.48 | 1.38 | -1.48 | 1.01 | -2.42 | 1.73 | 1.65 |
| antioxidant activity | 35 | 1.06 | 1.04 | -1.09 | -1.06 | -1.08 | -1.08 | 1.01 |
| ammonia ligase activity | 2 | 1.91 | -1.99 | 2.01 | -1.44 | 1.52 | -1.73 | -1.25 |
| kynurenine-oxoglutarate transaminase activity | 1 | 1.14 | 1.23 | -1.25 | 1.13 | -1.05 | 1.32 | 1.04 |
| acyl-CoA desaturase activity | 8 | -1.57 | -1.03 | -1.33 | 1.02 | -1.14 | -1.74 | 1.05 |
| procollagen-proline 4-dioxygenase complex | 17 | -1.04 | -1.0 | -1.12 | -1.02 | -1.15 | -1.03 | -1.07 |
| iron-sulfur cluster assembly | 7 | 1.2 | 1.16 | 1.26 | 1.15 | 1.02 | 1.28 | -1.07 |
| steroid dehydrogenase activity | 8 | 3.51 | -1.74 | -1.02 | -1.08 | -1.5 | -2.16 | -1.12 |
| sphingomyelin phosphodiesterase activator activity | 1 | -1.61 | 1.4 | 2.58 | 1.07 | 1.35 | 1.37 | 1.11 |
| beta-N-acetylglucosaminidase activity | 3 | 3.05 | -1.03 | -1.72 | -1.02 | 1.13 | -1.04 | 1.42 |
| HNK-1 sulfotransferase activity | 2 | 2.01 | 1.05 | 1.17 | 1.04 | 1.08 | 1.34 | -1.13 |
| telomere capping | 7 | 1.48 | 1.09 | 1.57 | 1.16 | 1.48 | 1.55 | 1.18 |
| macroautophagy | 6 | 1.33 | 1.14 | 1.38 | 1.2 | 1.17 | 1.27 | 1.01 |
| regulation of macroautophagy | 2 | -1.22 | 1.35 | 1.01 | 1.16 | 1.29 | 1.48 | -1.02 |
| negative regulation of macroautophagy | 1 | -1.06 | 1.16 | 1.2 | 1.34 | 1.78 | -1.33 | -1.11 |
| RNA interference | 28 | -1.01 | -1.0 | 1.16 | 1.07 | 1.32 | 1.18 | 1.02 |
| channel regulator activity | 8 | 1.18 | 1.06 | 1.08 | 1.02 | 1.08 | 1.11 | 1.04 |
| N-sulfoglucosamine sulfohydrolase activity | 1 | -1.48 | -1.35 | -1.19 | -1.31 | -1.2 | -1.3 | 1.23 |
| attachment of GPI anchor to protein | 2 | 1.49 | 1.08 | 1.21 | 1.27 | 1.12 | -1.05 | -1.02 |
| selenocysteine metabolic process | 2 | 1.76 | 1.25 | 1.23 | 1.04 | 1.04 | 1.62 | -1.07 |
| selenocysteine biosynthetic process | 2 | 1.76 | 1.25 | 1.23 | 1.04 | 1.04 | 1.62 | -1.07 |
| protein N-acetylglucosaminyltransferase activity | 3 | 1.03 | 1.05 | 1.06 | 1.08 | 1.08 | -1.0 | 1.16 |
| glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity | 1 | -3.7 | 1.32 | 1.71 | 1.03 | 1.54 | -3.12 | 1.7 |
| death | 156 | 1.07 | 1.04 | 1.16 | 1.06 | 1.25 | 1.12 | 1.15 |
| O-glycan processing | 3 | -2.23 | 1.15 | 1.26 | 1.19 | 1.31 | -2.5 | 1.12 |
| O-glycan processing, core 1 | 1 | -3.7 | 1.32 | 1.71 | 1.03 | 1.54 | -3.12 | 1.7 |
| tissue death | 79 | -1.03 | 1.02 | 1.13 | 1.07 | 1.24 | 1.12 | 1.1 |
| prefoldin complex | 10 | 1.4 | -1.02 | 1.21 | 1.29 | 1.23 | 1.13 | -1.24 |
| arginine N-methyltransferase activity | 9 | 1.19 | 1.07 | 1.15 | 1.17 | 1.1 | 1.02 | 1.01 |
| protein-arginine N-methyltransferase activity | 9 | 1.19 | 1.07 | 1.15 | 1.17 | 1.1 | 1.02 | 1.01 |
| lysine N-methyltransferase activity | 14 | 1.29 | 1.27 | 1.5 | 1.07 | 1.4 | 1.43 | 1.38 |
| protein-lysine N-methyltransferase activity | 14 | 1.29 | 1.27 | 1.5 | 1.07 | 1.4 | 1.43 | 1.38 |
| eukaryotic 43S preinitiation complex | 1 | 2.17 | 1.18 | 1.52 | -1.1 | 1.33 | 2.16 | 1.32 |
| small conductance calcium-activated potassium channel activity | 1 | 1.12 | -1.04 | 1.03 | 1.09 | -1.03 | 1.27 | -1.03 |
| glycerone-phosphate O-acyltransferase activity | 1 | 2.26 | 1.05 | 1.14 | 1.04 | 1.53 | -1.65 | 1.6 |
| CoA hydrolase activity | 4 | 1.3 | 1.08 | 1.01 | 1.04 | -1.02 | 1.91 | -1.09 |
| myristoyl-[acyl-carrier-protein] hydrolase activity | 2 | 3.49 | -1.07 | -1.08 | -1.15 | 1.4 | 1.97 | 2.1 |
| palmitoyl-[acyl-carrier-protein] hydrolase activity | 2 | 3.49 | -1.07 | -1.08 | -1.15 | 1.4 | 1.97 | 2.1 |
| acyl-[acyl-carrier-protein] hydrolase activity | 2 | 3.49 | -1.07 | -1.08 | -1.15 | 1.4 | 1.97 | 2.1 |
| lipase activity | 84 | -1.92 | -1.25 | -1.55 | -1.09 | -1.32 | -2.03 | -1.49 |
| negative regulation of G-protein coupled receptor protein signaling pathway | 11 | 1.43 | 1.12 | 1.43 | 1.14 | 1.25 | 1.34 | 1.24 |
| kinase activity | 351 | 1.18 | 1.1 | 1.21 | 1.06 | 1.23 | 1.3 | 1.12 |
| phosphorylation | 353 | 1.17 | 1.09 | 1.23 | 1.06 | 1.23 | 1.4 | 1.08 |
| dephosphorylation | 108 | 1.34 | 1.06 | 1.15 | 1.03 | 1.19 | 1.32 | 1.14 |
| inositol bisphosphate phosphatase activity | 1 | 3.45 | -1.15 | 1.32 | 1.0 | -1.07 | 1.6 | -1.16 |
| phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| ommatidial rotation | 26 | 1.05 | 1.09 | 1.3 | 1.04 | 1.44 | 1.22 | 1.4 |
| mushroom body development | 51 | 1.06 | 1.12 | 1.3 | 1.08 | 1.25 | 1.26 | 1.19 |
| endoplasmic reticulum membrane fusion | 2 | 1.13 | 1.31 | -1.2 | 1.21 | 1.07 | -1.49 | 1.37 |
| female meiosis chromosome segregation | 30 | 1.12 | 1.08 | 1.2 | 1.12 | 1.26 | 1.19 | 1.16 |
| neuron remodeling | 18 | 1.16 | 1.02 | 1.26 | 1.04 | 1.45 | 1.47 | 1.09 |
| basolateral plasma membrane | 34 | 1.42 | 1.0 | 1.32 | -1.08 | 1.36 | 1.58 | 1.36 |
| apical plasma membrane | 27 | 1.33 | 1.11 | 1.29 | 1.04 | 1.28 | 1.55 | 1.44 |
| oocyte microtubule cytoskeleton organization | 31 | 1.08 | 1.2 | 1.51 | 1.08 | 1.36 | 1.59 | 1.25 |
| apicolateral plasma membrane | 50 | 1.77 | -1.05 | 1.29 | -1.11 | 1.27 | 2.3 | 1.5 |
| lateral plasma membrane | 9 | 2.47 | -1.11 | 1.23 | -1.25 | 1.22 | 2.79 | 1.7 |
| regulation of apoptotic process | 92 | 1.16 | 1.13 | 1.27 | 1.08 | 1.29 | 1.25 | 1.23 |
| second mitotic wave involved in compound eye morphogenesis | 8 | 1.1 | 1.21 | 1.15 | 1.19 | 1.36 | 1.3 | 1.21 |
| morphogenesis of embryonic epithelium | 105 | 1.16 | 1.08 | 1.34 | 1.03 | 1.31 | 1.33 | 1.31 |
| establishment or maintenance of polarity of embryonic epithelium | 8 | 1.29 | 1.17 | 1.66 | 1.02 | 1.38 | 1.51 | 1.32 |
| morphogenesis of follicular epithelium | 30 | 1.19 | 1.16 | 1.46 | 1.13 | 1.44 | 1.35 | 1.23 |
| establishment or maintenance of polarity of follicular epithelium | 20 | 1.28 | 1.14 | 1.48 | 1.13 | 1.41 | 1.28 | 1.19 |
| morphogenesis of larval imaginal disc epithelium | 8 | 1.12 | 1.06 | 1.78 | 1.05 | 1.53 | 1.28 | 1.0 |
| establishment or maintenance of polarity of larval imaginal disc epithelium | 4 | 1.09 | 1.11 | 2.19 | -1.04 | 1.64 | 1.48 | 1.2 |
| cell-cell adhesion | 54 | 1.19 | -1.01 | 1.06 | -1.05 | 1.29 | 1.29 | 1.35 |
| calcium-dependent cell-matrix adhesion | 2 | -3.06 | 1.2 | 1.48 | 1.02 | 1.55 | -3.22 | 1.1 |
| catenin complex | 2 | 1.43 | 1.06 | 1.57 | 1.11 | 1.6 | 1.7 | 1.6 |
| meiotic chromosome movement towards spindle pole | 1 | 1.13 | 1.05 | -1.04 | 1.47 | 1.34 | -1.18 | -1.11 |
| male meiotic chromosome movement towards spindle pole | 1 | 1.13 | 1.05 | -1.04 | 1.47 | 1.34 | -1.18 | -1.11 |
| imaginal disc-derived leg joint morphogenesis | 8 | -2.19 | 1.03 | -1.69 | -1.64 | 2.1 | 1.1 | 1.32 |
| dendrite development | 132 | 1.18 | 1.12 | 1.28 | 1.11 | 1.28 | 1.22 | 1.16 |
| sensory organ precursor cell fate determination | 19 | 1.13 | 1.05 | 1.17 | 1.04 | 1.28 | 1.16 | 1.21 |
| activin receptor activity, type I | 1 | -1.85 | 1.82 | 2.01 | 1.27 | 1.8 | 1.64 | 1.57 |
| palmitoyl-CoA oxidase activity | 3 | 1.38 | -1.55 | -1.39 | 1.44 | 1.71 | -2.77 | -1.43 |
| pristanoyl-CoA oxidase activity | 2 | -15.88 | 1.87 | -3.13 | -1.51 | -1.49 | -10.5 | -1.82 |
| dimethylargininase activity | 1 | 1.77 | 1.6 | 1.45 | 1.13 | 1.23 | 1.13 | 1.16 |
| CoA-ligase activity | 13 | -2.56 | -1.1 | -1.57 | -1.26 | -1.84 | -1.46 | -2.12 |
| carnitine O-acyltransferase activity | 5 | 1.08 | 1.06 | -1.39 | 1.16 | 1.37 | -1.36 | -1.1 |
| acetyltransferase activity | 54 | -1.01 | 1.14 | 1.0 | 1.14 | 1.18 | -1.11 | 1.03 |
| C-acyltransferase activity | 8 | 1.16 | 1.02 | 1.03 | 1.15 | 1.21 | -1.21 | -1.3 |
| palmitoyltransferase activity | 25 | -1.15 | 1.11 | 1.19 | 1.13 | 1.12 | -1.19 | -1.11 |
| N-acyltransferase activity | 45 | -1.1 | 1.17 | 1.02 | 1.17 | 1.18 | -1.19 | 1.02 |
| acylglycerol O-acyltransferase activity | 7 | -1.48 | -1.12 | 1.19 | 1.18 | 2.2 | -2.06 | -1.75 |
| O-acetyltransferase activity | 5 | 1.07 | 1.05 | -1.14 | 1.08 | 1.21 | -1.26 | -1.0 |
| O-octanoyltransferase activity | 1 | 2.33 | -1.31 | -2.71 | 1.32 | 2.24 | -2.61 | 1.02 |
| octanoyltransferase activity | 2 | 2.01 | -1.16 | -1.62 | 1.18 | 1.39 | -1.12 | -1.11 |
| O-palmitoyltransferase activity | 1 | 1.55 | 1.41 | -1.09 | 1.13 | 1.34 | -1.06 | 1.05 |
| S-acyltransferase activity | 27 | 1.03 | 1.08 | 1.14 | 1.06 | 1.1 | 1.04 | -1.05 |
| S-acetyltransferase activity | 3 | 2.46 | 1.01 | 1.19 | -1.13 | 1.38 | 2.35 | 1.7 |
| S-malonyltransferase activity | 3 | 2.14 | -1.05 | -1.1 | -1.07 | 1.17 | 1.47 | 1.42 |
| malonyltransferase activity | 3 | 2.14 | -1.05 | -1.1 | -1.07 | 1.17 | 1.47 | 1.42 |
| CoA carboxylase activity | 3 | 2.45 | 1.39 | 1.08 | -1.18 | 1.31 | 2.01 | 1.86 |
| mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity | 2 | 1.39 | 1.18 | 1.42 | 1.45 | 1.47 | 1.2 | 1.02 |
| tRNA (guanine) methyltransferase activity | 2 | 1.37 | 1.35 | 1.16 | 1.36 | 1.21 | 1.09 | 1.14 |
| tRNA (adenine) methyltransferase activity | 1 | 1.03 | 1.07 | -1.39 | 1.43 | 1.1 | -1.31 | -1.42 |
| tRNA (cytosine) methyltransferase activity | 1 | -1.06 | 1.07 | 1.05 | 1.57 | 1.09 | -1.37 | -1.08 |
| tRNA (cytosine-5-)-methyltransferase activity | 1 | -1.06 | 1.07 | 1.05 | 1.57 | 1.09 | -1.37 | -1.08 |
| tRNA (adenine-N1-)-methyltransferase activity | 1 | 1.03 | 1.07 | -1.39 | 1.43 | 1.1 | -1.31 | -1.42 |
| rRNA (adenine) methyltransferase activity | 2 | 1.61 | 1.27 | 1.44 | 1.22 | 1.57 | 1.46 | 1.13 |
| rRNA (uridine) methyltransferase activity | 3 | 1.55 | 1.21 | 1.1 | 1.32 | 1.19 | 1.02 | -1.14 |
| posttranscriptional gene silencing | 37 | 1.12 | 1.04 | 1.18 | 1.1 | 1.27 | 1.33 | 1.07 |
| RNA-induced silencing complex | 7 | -1.01 | 1.19 | 1.52 | 1.03 | 1.34 | 1.15 | 1.36 |
| bidentate ribonuclease III activity | 1 | -1.14 | 1.08 | 1.42 | -1.27 | 1.25 | 1.05 | 1.3 |
| somatic cell DNA recombination | 1 | -1.03 | 1.26 | 1.39 | 1.47 | 1.41 | -1.54 | -1.18 |
| C-acetyltransferase activity | 3 | 1.18 | 1.04 | -1.11 | 1.03 | -1.14 | 1.32 | -1.35 |
| C-palmitoyltransferase activity | 2 | -2.24 | 1.07 | 2.48 | 1.17 | 1.31 | -1.71 | -1.18 |
| X chromosome located dosage compensation complex, transcription activating | 6 | 1.37 | 1.23 | 1.41 | 1.11 | 1.23 | 1.51 | 1.37 |
| dosage compensation complex assembly involved in dosage compensation by hyperactivation of X chromosome | 3 | 1.47 | 1.24 | 1.5 | 1.08 | 1.32 | 1.62 | 1.47 |
| gene silencing | 82 | 1.19 | 1.1 | 1.25 | 1.12 | 1.3 | 1.29 | 1.13 |
| myosin complex | 26 | -1.12 | 1.01 | 1.34 | -1.05 | 1.23 | 1.03 | 1.01 |
| myosin II complex | 7 | 2.46 | -1.39 | 1.28 | -1.33 | 1.16 | 2.04 | -1.08 |
| unconventional myosin complex | 8 | -1.34 | 1.12 | 1.71 | 1.02 | 1.43 | -1.05 | 1.11 |
| pyrophosphatase activity | 545 | 1.04 | 1.07 | 1.18 | 1.06 | 1.17 | 1.12 | 1.03 |
| vacuolar proton-transporting V-type ATPase complex | 25 | -1.7 | 1.0 | 1.39 | -1.2 | -1.2 | 1.32 | -1.15 |
| regulation of embryonic cell shape | 30 | 1.15 | 1.18 | 1.42 | 1.07 | 1.45 | 1.47 | 1.31 |
| cell migration | 188 | 1.14 | 1.09 | 1.33 | 1.05 | 1.37 | 1.23 | 1.22 |
| negative regulation of transcription from RNA polymerase III promoter | 2 | 2.38 | 1.05 | 1.63 | 1.37 | 1.48 | 1.65 | 1.1 |
| cytoplasmic transport | 18 | -1.19 | 1.13 | 1.22 | 1.09 | 1.2 | -1.37 | -1.03 |
| tryptophan hydroxylase activator activity | 1 | 2.28 | 1.19 | 1.07 | -1.0 | 1.37 | 2.2 | 2.5 |
| peptidase activator activity | 8 | 1.1 | 1.12 | 1.28 | 1.11 | 1.16 | -1.1 | 1.31 |
| peptide hormone processing | 2 | -3.69 | 1.49 | 2.0 | -1.05 | 1.12 | -3.53 | 1.27 |
| structural constituent of peritrophic membrane | 24 | -1.99 | -1.49 | -1.95 | -1.18 | -1.6 | -2.02 | 1.66 |
| protein-hormone receptor activity | 6 | 1.04 | -1.23 | -1.1 | -1.21 | -1.12 | 1.72 | -1.28 |
| pheromone receptor activity | 1 | 1.02 | 1.0 | -1.07 | 1.11 | 1.03 | 1.03 | -1.03 |
| apoptotic protease activator activity | 3 | 1.67 | 1.11 | 1.24 | 1.1 | 1.19 | 1.2 | 1.4 |
| mitochondrial fatty acid beta-oxidation multienzyme complex | 2 | 2.42 | 1.07 | -1.08 | 1.02 | 1.26 | 1.66 | -1.08 |
| long-chain-enoyl-CoA hydratase activity | 1 | 2.19 | 1.13 | 1.21 | -1.05 | 1.43 | 2.22 | 1.16 |
| long-chain-3-hydroxyacyl-CoA dehydrogenase activity | 2 | 2.42 | 1.07 | -1.08 | 1.02 | 1.26 | 1.66 | -1.08 |
| core-binding factor complex | 2 | 1.8 | -1.02 | -1.19 | 1.07 | -1.13 | 1.06 | -1.05 |
| growth hormone-releasing hormone receptor activity | 3 | 1.44 | -1.06 | -1.09 | -1.03 | -1.2 | 1.08 | -1.08 |
| latrotoxin receptor activity | 1 | -1.05 | -1.14 | -1.15 | 1.03 | 1.21 | 1.62 | 2.05 |
| sarcoplasm | 4 | 2.04 | -1.34 | 1.12 | -1.41 | -1.15 | -1.13 | 1.11 |
| sarcoplasmic reticulum | 3 | 2.05 | -1.41 | 1.18 | -1.55 | -1.18 | -1.27 | 1.12 |
| metallochaperone activity | 3 | 1.48 | 1.1 | 1.32 | 1.12 | 1.03 | 1.6 | -1.13 |
| microvillus assembly | 1 | -2.22 | -1.56 | -1.05 | -1.91 | -2.01 | -2.21 | 7.0 |
| copper chaperone activity | 2 | 1.54 | 1.13 | 1.31 | -1.02 | -1.15 | 1.51 | -1.05 |
| superoxide dismutase copper chaperone activity | 1 | 1.35 | 1.23 | -1.1 | 1.09 | -1.09 | -1.29 | 1.14 |
| cyclin-dependent protein kinase 5 holoenzyme complex | 2 | -1.11 | 1.16 | 1.23 | 1.19 | 1.1 | -1.28 | -1.16 |
| cyclin-dependent protein kinase 5 activator activity | 1 | 1.11 | 1.03 | 1.17 | 1.14 | 1.15 | -1.04 | -1.04 |
| cyclin-dependent protein kinase regulator activity | 19 | 1.26 | 1.15 | 1.26 | 1.19 | 1.43 | 1.03 | 1.11 |
| intein-mediated protein splicing | 1 | -1.72 | -1.51 | 1.07 | 1.74 | 1.86 | -2.25 | -3.98 |
| protein autoprocessing | 4 | 1.71 | -1.09 | 1.55 | 1.25 | 1.59 | 1.29 | -1.21 |
| male courtship behavior, orientation prior to leg tapping and wing vibration | 2 | -1.07 | 1.03 | 1.04 | -1.02 | -1.04 | -1.08 | 1.02 |
| male courtship behavior, veined wing vibration | 13 | 1.07 | 1.04 | 1.44 | 1.14 | 1.09 | -1.01 | 1.01 |
| male courtship behavior, proboscis-mediated licking | 1 | 1.27 | 1.02 | 1.28 | 1.01 | 1.16 | 1.03 | 1.05 |
| base conversion or substitution editing | 1 | 1.13 | 1.14 | 1.55 | 1.01 | 1.4 | 2.0 | 1.26 |
| mRNA modification | 1 | 1.13 | 1.14 | 1.55 | 1.01 | 1.4 | 2.0 | 1.26 |
| protein import into peroxisome matrix | 1 | -1.21 | 1.08 | -2.37 | 1.65 | 1.29 | -3.85 | -2.28 |
| peroxisome fission | 3 | 2.08 | -1.17 | -1.4 | 1.43 | 1.51 | -1.91 | 1.09 |
| protein import into peroxisome matrix, docking | 1 | -1.21 | 1.08 | -2.37 | 1.65 | 1.29 | -3.85 | -2.28 |
| positive regulation of transcription, DNA-dependent | 201 | 1.12 | 1.04 | 1.13 | 1.04 | 1.13 | 1.19 | 1.15 |
| negative regulation of transcription, DNA-dependent | 184 | 1.17 | 1.08 | 1.16 | 1.04 | 1.22 | 1.22 | 1.16 |
| chromatin modification | 91 | 1.23 | 1.19 | 1.32 | 1.15 | 1.35 | 1.32 | 1.23 |
| covalent chromatin modification | 63 | 1.22 | 1.17 | 1.31 | 1.16 | 1.35 | 1.33 | 1.21 |
| histone modification | 63 | 1.22 | 1.17 | 1.31 | 1.16 | 1.35 | 1.33 | 1.21 |
| histone methylation | 26 | 1.36 | 1.2 | 1.41 | 1.14 | 1.42 | 1.43 | 1.28 |
| histone phosphorylation | 7 | -1.05 | 1.06 | 1.11 | 1.18 | 1.26 | -1.01 | 1.01 |
| histone acetylation | 14 | 1.4 | 1.24 | 1.42 | 1.18 | 1.37 | 1.44 | 1.29 |
| histone deacetylation | 5 | 1.19 | 1.12 | 1.01 | 1.23 | 1.22 | 1.44 | 1.15 |
| histone demethylation | 7 | 1.18 | 1.25 | 1.72 | 1.13 | 1.59 | 1.68 | 1.3 |
| histone deubiquitination | 5 | 1.17 | 1.1 | 1.27 | 1.13 | 1.23 | 1.11 | 1.2 |
| protein deubiquitination | 12 | 1.11 | 1.1 | 1.41 | 1.13 | 1.34 | 1.22 | 1.21 |
| Sin3 complex | 3 | 1.26 | 1.15 | 1.58 | 1.16 | 1.67 | 1.6 | 1.21 |
| NuRD complex | 4 | 1.58 | 1.23 | 1.59 | 1.18 | 1.53 | 1.55 | 1.45 |
| non-covalent chromatin modification | 1 | 1.45 | 1.01 | 1.08 | 1.38 | 1.13 | -1.44 | -1.23 |
| nucleosome positioning | 3 | 1.19 | 1.07 | 1.82 | 1.21 | 1.88 | 1.74 | 1.38 |
| chromatin remodeling complex | 70 | 1.23 | 1.14 | 1.26 | 1.2 | 1.32 | 1.19 | 1.11 |
| NURF complex | 5 | 1.15 | 1.22 | 1.63 | 1.19 | 1.59 | 1.56 | 1.32 |
| ACF complex | 5 | 1.33 | -1.77 | -1.21 | -1.79 | 1.3 | 2.26 | 1.17 |
| DNA-directed RNA polymerase II, holoenzyme | 51 | 1.34 | 1.12 | 1.26 | 1.2 | 1.28 | 1.16 | 1.09 |
| mediator complex | 32 | 1.41 | 1.08 | 1.29 | 1.24 | 1.33 | 1.23 | 1.06 |
| Cdc73/Paf1 complex | 2 | -1.06 | 1.05 | 1.02 | 1.24 | 1.1 | 1.21 | -1.3 |
| amino acid binding | 15 | -1.42 | -1.07 | -1.49 | 1.17 | 1.15 | -1.1 | -1.49 |
| protein arginylation | 1 | 1.03 | 1.39 | 1.46 | 1.24 | 1.37 | 1.69 | 1.36 |
| flotillin complex | 1 | 3.68 | -1.06 | -1.03 | 1.16 | 1.0 | 1.99 | -1.04 |
| Rac protein signal transduction | 2 | -1.38 | 1.43 | 1.79 | 1.19 | 1.71 | -1.05 | 1.42 |
| CCAAT-binding factor complex | 1 | 4.06 | 1.06 | 1.04 | 1.34 | 1.6 | 1.05 | 1.52 |
| glutaminyl-peptide cyclotransferase activity | 2 | 2.71 | -1.35 | -1.37 | -1.7 | -1.26 | -1.18 | 2.94 |
| nuclear body | 27 | -1.05 | 1.08 | 1.11 | 1.12 | 1.17 | 1.02 | -1.04 |
| nuclear speck | 12 | -1.25 | 1.18 | 1.12 | 1.11 | 1.16 | -1.02 | -1.07 |
| oxidoreductase activity, acting on CH-OH group of donors | 112 | 1.18 | -1.01 | -1.4 | -1.09 | -1.09 | 1.09 | 1.07 |
| malate dehydrogenase activity | 6 | -1.07 | 1.16 | 1.1 | -1.34 | -1.06 | 1.05 | 1.38 |
| oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 74 | 1.28 | 1.01 | -1.37 | -1.14 | -1.09 | 1.12 | 1.17 |
| malate dehydrogenase (oxaloacetate-decarboxylating) activity | 4 | -1.07 | 1.26 | 1.14 | -1.52 | -1.06 | 1.05 | 1.65 |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | 34 | 1.38 | -1.54 | -1.84 | -1.66 | -1.29 | 1.37 | 1.27 |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors, cytochrome as acceptor | 1 | 2.72 | -3.32 | -2.57 | -3.03 | -1.93 | 22.15 | -2.83 |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor | 7 | -1.14 | 1.15 | 1.33 | 1.02 | 1.18 | 1.45 | -1.19 |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors | 42 | 1.3 | -1.42 | -1.6 | -1.54 | -1.21 | 1.48 | 1.15 |
| oxidoreductase activity, acting on the CH-CH group of donors | 38 | 1.11 | 1.12 | -1.03 | 1.1 | 1.11 | -1.16 | -1.13 |
| oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | 5 | 1.34 | 1.13 | 1.06 | 1.07 | 1.1 | 1.13 | 1.09 |
| oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor | 11 | -1.64 | 1.16 | -1.22 | 1.19 | 1.14 | -2.19 | -1.33 |
| oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor | 6 | -1.06 | 1.13 | 1.29 | 1.05 | -1.01 | 1.24 | -1.27 |
| oxidoreductase activity, acting on the CH-NH2 group of donors | 13 | 1.11 | -1.17 | -1.04 | 1.09 | 1.13 | 1.04 | -1.25 |
| oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor | 3 | 1.19 | -1.14 | 1.18 | -1.25 | 1.27 | 2.68 | -1.22 |
| oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor | 8 | -1.02 | -1.07 | -1.3 | 1.32 | 1.19 | -1.15 | -1.32 |
| oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor | 1 | -1.37 | -1.08 | -1.16 | -1.21 | -1.14 | 1.68 | -1.06 |
| oxidoreductase activity, acting on the CH-NH group of donors | 20 | 1.39 | -1.16 | -1.24 | -1.21 | -1.13 | 1.8 | -1.06 |
| oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor | 13 | 1.38 | -1.17 | -1.24 | -1.18 | -1.19 | 1.9 | -1.08 |
| transferase activity, transferring amino-acyl groups | 10 | 1.86 | -1.26 | -1.16 | -1.23 | 1.18 | 1.3 | 1.33 |
| oxidoreductase activity, acting on the CH-NH group of donors, disulfide as acceptor | 4 | 1.29 | 1.17 | 1.1 | 1.04 | 1.18 | -1.03 | 1.04 |
| oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor | 1 | 2.09 | 1.59 | -1.41 | 1.19 | 1.36 | 1.02 | -1.19 |
| oxidoreductase activity, acting on NADH or NADPH, heme protein as acceptor | 2 | 1.22 | 1.18 | 1.02 | 1.17 | 1.12 | -1.17 | 1.16 |
| oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor | 30 | 1.12 | 1.03 | 1.27 | 1.07 | 1.1 | 1.73 | -1.3 |
| oxidoreductase activity, acting on NADH or NADPH, nitrogenous group as acceptor | 1 | 1.29 | 1.1 | 1.03 | 1.22 | 1.29 | 1.66 | -1.14 |
| oxidoreductase activity, acting on other nitrogenous compounds as donors | 2 | 2.03 | -1.43 | -1.4 | -1.33 | -1.08 | 1.05 | -1.28 |
| oxidoreductase activity, acting on other nitrogenous compounds as donors, oxygen as acceptor | 1 | 3.87 | -2.08 | -2.11 | -2.0 | -1.26 | -1.05 | -1.64 |
| oxidoreductase activity, acting on a sulfur group of donors | 43 | 1.04 | 1.02 | -1.07 | 1.17 | 1.05 | -1.05 | -1.15 |
| oxidoreductase activity, acting on a sulfur group of donors, NAD or NADP as acceptor | 3 | -1.35 | 1.14 | 1.07 | -1.01 | -1.07 | 1.01 | -1.04 |
| oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor | 6 | -1.13 | 1.15 | 1.03 | 1.17 | 1.02 | 1.31 | -1.14 |
| oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor | 3 | -1.6 | -1.4 | -2.5 | 1.38 | -1.37 | -2.12 | -1.83 |
| oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor | 4 | 1.29 | 1.17 | 1.1 | 1.04 | 1.18 | -1.03 | 1.04 |
| oxidoreductase activity, acting on a heme group of donors | 18 | -1.12 | -1.08 | 1.07 | -1.07 | 1.03 | 1.19 | -1.31 |
| oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor | 18 | -1.12 | -1.08 | 1.07 | -1.07 | 1.03 | 1.19 | -1.31 |
| oxidoreductase activity, acting on diphenols and related substances as donors | 14 | -1.21 | -1.11 | -1.11 | -1.13 | -1.22 | 1.0 | -1.09 |
| oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor | 10 | -1.05 | 1.06 | 1.14 | 1.09 | 1.02 | 1.34 | -1.41 |
| oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor | 4 | -1.73 | -1.69 | -2.0 | -1.92 | -2.12 | -2.06 | 1.73 |
| oxidoreductase activity, acting on peroxide as acceptor | 22 | -1.07 | -1.02 | -1.12 | -1.17 | -1.19 | -1.23 | 1.01 |
| oxidoreductase activity, acting on single donors with incorporation of molecular oxygen | 34 | 1.11 | -1.05 | -1.11 | -1.04 | -1.12 | 1.08 | -1.07 |
| oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | 33 | 1.07 | -1.01 | -1.07 | -1.0 | -1.09 | 1.03 | -1.05 |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 168 | -1.09 | -1.09 | -1.25 | -1.02 | -1.03 | -1.11 | -1.07 |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 45 | -1.05 | 1.04 | -1.04 | 1.03 | 1.0 | 1.02 | -1.01 |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NADH or NADPH as one donor, and incorporation of one atom of oxygen | 7 | 1.08 | -1.49 | -2.16 | -1.08 | -1.58 | -1.06 | -1.04 |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen | 1 | 1.6 | 1.35 | -1.72 | -1.6 | -1.17 | -1.33 | 1.5 |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen | 3 | 1.14 | -1.23 | -1.27 | -1.21 | -1.15 | 1.59 | -1.01 |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen | 4 | 1.53 | 1.13 | 1.14 | 1.01 | 1.23 | 1.29 | 1.28 |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen | 3 | 1.29 | 1.11 | -1.09 | -1.0 | -1.13 | -1.11 | -1.01 |
| oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water | 10 | -1.69 | -1.14 | -1.69 | -1.05 | -1.38 | -1.35 | 1.04 |
| oxidoreductase activity, acting on superoxide radicals as acceptor | 5 | 1.45 | 1.15 | -1.15 | 1.12 | -1.07 | 1.37 | -1.06 |
| oxidoreductase activity, oxidizing metal ions | 4 | -1.43 | -2.04 | -1.83 | -1.64 | 1.18 | -1.2 | 1.72 |
| oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor | 1 | 1.59 | 1.02 | 1.27 | 1.16 | 1.58 | 1.37 | 1.04 |
| oxidoreductase activity, oxidizing metal ions, oxygen as acceptor | 3 | -1.88 | -2.6 | -2.43 | -2.04 | 1.07 | -1.41 | 2.03 |
| oxidoreductase activity, acting on CH or CH2 groups | 4 | -1.34 | 1.29 | 1.11 | 1.02 | 1.25 | -1.45 | 1.04 |
| oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor | 2 | 1.25 | 1.78 | -1.17 | -1.32 | -1.19 | -1.3 | 1.66 |
| oxidoreductase activity, acting on CH or CH2 groups, oxygen as acceptor | 1 | 1.38 | 1.39 | -1.09 | 1.59 | 1.39 | -1.89 | 1.34 |
| oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor | 2 | -2.27 | -1.06 | 1.44 | 1.36 | 1.87 | -1.61 | -1.54 |
| oxidoreductase activity, acting on iron-sulfur proteins as donors | 2 | -1.02 | 1.13 | 1.16 | 1.22 | 1.24 | -1.06 | -1.06 |
| oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor | 2 | -1.02 | 1.13 | 1.16 | 1.22 | 1.24 | -1.06 | -1.06 |
| transferase activity | 1025 | 1.03 | 1.07 | 1.07 | 1.05 | 1.15 | 1.03 | 1.07 |
| transferase activity, transferring one-carbon groups | 103 | 1.29 | 1.11 | 1.16 | 1.16 | 1.2 | 1.21 | 1.03 |
| hydroxymethyl-, formyl- and related transferase activity | 5 | 1.33 | 1.06 | 1.25 | 1.15 | 1.17 | 1.45 | -1.08 |
| carboxyl- or carbamoyltransferase activity | 1 | -1.12 | 1.14 | 1.63 | 1.21 | 1.33 | 1.0 | 1.05 |
| transferase activity, transferring aldehyde or ketonic groups | 3 | 2.36 | 1.15 | -2.0 | 1.02 | -1.05 | 1.23 | 1.57 |
| transferase activity, transferring acyl groups | 158 | -1.13 | 1.02 | -1.11 | 1.07 | 1.11 | -1.28 | -1.14 |
| transferase activity, transferring acyl groups other than amino-acyl groups | 137 | -1.17 | 1.03 | -1.11 | 1.09 | 1.1 | -1.33 | -1.17 |
| succinyltransferase activity | 2 | 1.41 | 1.15 | 1.17 | -1.16 | -1.1 | 2.22 | -1.06 |
| N-succinyltransferase activity | 1 | 1.58 | 1.36 | -1.31 | -1.09 | -1.26 | 1.25 | 1.23 |
| S-succinyltransferase activity | 1 | 1.27 | -1.02 | 1.79 | -1.24 | 1.05 | 3.95 | -1.37 |
| myosin I binding | 1 | -1.18 | 1.13 | 1.19 | 1.15 | 1.38 | -1.04 | 1.11 |
| transferase activity, transferring glycosyl groups | 167 | -1.11 | -1.01 | -1.01 | 1.01 | 1.1 | -1.1 | 1.15 |
| transferase activity, transferring hexosyl groups | 139 | -1.18 | -1.01 | -1.03 | 1.01 | 1.11 | -1.14 | 1.16 |
| transferase activity, transferring pentosyl groups | 19 | 1.06 | 1.01 | 1.09 | -1.01 | -1.01 | -1.15 | 1.07 |
| transferase activity, transferring alkyl or aryl (other than methyl) groups | 58 | -1.11 | 1.06 | 1.05 | -1.09 | 1.02 | -1.25 | 1.03 |
| spermine synthase activity | 1 | -1.46 | 1.37 | 1.59 | 1.23 | 1.4 | 2.11 | 1.08 |
| transferase activity, transferring nitrogenous groups | 19 | -1.08 | 1.17 | -1.23 | -1.08 | -1.02 | -1.02 | 1.19 |
| transferase activity, transferring phosphorus-containing groups | 487 | 1.08 | 1.11 | 1.15 | 1.05 | 1.2 | 1.16 | 1.12 |
| phosphotransferase activity, alcohol group as acceptor | 326 | 1.17 | 1.11 | 1.21 | 1.07 | 1.25 | 1.31 | 1.14 |
| phosphotransferase activity, carboxyl group as acceptor | 3 | -2.35 | 1.21 | -1.51 | -1.01 | -1.08 | -1.33 | 1.07 |
| phosphotransferase activity, nitrogenous group as acceptor | 6 | 1.06 | -1.03 | 1.15 | -1.01 | 1.23 | 1.37 | -1.18 |
| phosphotransferase activity, phosphate group as acceptor | 19 | 1.61 | -1.01 | 1.22 | 1.02 | 1.17 | 1.37 | -1.03 |
| diphosphotransferase activity | 4 | 1.52 | 1.16 | 1.32 | 1.12 | 1.12 | 1.57 | 1.12 |
| nucleotidyltransferase activity | 70 | 1.17 | 1.12 | 1.21 | 1.13 | 1.2 | 1.05 | 1.07 |
| phosphotransferase activity, for other substituted phosphate groups | 9 | -1.26 | 1.22 | 1.5 | 1.13 | 1.37 | -1.13 | 1.06 |
| phosphotransferase activity, paired acceptors | 2 | 1.76 | 1.25 | 1.23 | 1.04 | 1.04 | 1.62 | -1.07 |
| transferase activity, transferring sulfur-containing groups | 28 | 1.25 | 1.07 | 1.14 | 1.01 | 1.07 | 1.02 | 1.1 |
| transferase activity, transferring selenium-containing groups | 1 | 6.68 | -1.68 | 1.43 | -1.05 | 1.36 | 2.98 | 1.28 |
| hydrolase activity | 1780 | -1.23 | -1.08 | -1.14 | -1.01 | -1.01 | -1.21 | -1.14 |
| hydrolase activity, acting on ester bonds | 445 | 1.01 | -1.0 | -1.06 | 1.01 | 1.06 | -1.01 | -1.04 |
| thiolester hydrolase activity | 39 | 1.28 | 1.18 | 1.36 | 1.14 | 1.33 | 1.38 | 1.2 |
| phosphatase activity | 173 | 1.11 | 1.02 | -1.05 | 1.0 | 1.12 | 1.12 | 1.01 |
| diphosphoric monoester hydrolase activity | 1 | 1.81 | -1.03 | 1.57 | 1.17 | 1.71 | 1.48 | -1.14 |
| exonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters | 24 | 1.21 | 1.16 | 1.16 | 1.21 | 1.26 | 1.12 | 1.05 |
| hydrolase activity, acting on glycosyl bonds | 100 | -1.69 | -1.27 | -2.25 | -1.21 | -1.43 | -1.66 | -1.24 |
| hydrolase activity, hydrolyzing N-glycosyl compounds | 6 | 2.07 | -1.07 | 1.03 | -1.04 | 1.05 | 1.76 | 1.02 |
| hydrolase activity, acting on ether bonds | 7 | -2.36 | -1.14 | -1.27 | 1.0 | -1.04 | -1.57 | 1.01 |
| trialkylsulfonium hydrolase activity | 3 | -2.93 | -1.82 | -1.21 | -1.07 | 1.22 | 1.86 | -1.16 |
| ether hydrolase activity | 4 | -2.01 | 1.25 | -1.31 | 1.06 | -1.24 | -3.52 | 1.14 |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 85 | -1.06 | -1.12 | -1.21 | -1.01 | -1.05 | -1.02 | -1.1 |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides | 51 | -1.18 | -1.24 | -1.25 | 1.0 | -1.05 | -1.18 | -1.25 |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides | 4 | 1.19 | 1.06 | 1.03 | 1.24 | 1.04 | -1.03 | -1.09 |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines | 3 | 1.74 | 1.1 | 1.15 | 1.01 | -1.04 | 1.8 | 1.17 |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines | 20 | 1.0 | 1.02 | -1.24 | -1.12 | -1.14 | 1.23 | 1.04 |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in nitriles | 2 | 1.33 | 1.42 | -1.12 | 1.25 | 1.47 | 1.28 | -1.05 |
| hydrolase activity, acting on acid anhydrides | 553 | 1.04 | 1.07 | 1.17 | 1.06 | 1.16 | 1.12 | 1.03 |
| hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides | 551 | 1.04 | 1.07 | 1.17 | 1.06 | 1.16 | 1.12 | 1.03 |
| hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | 120 | -1.08 | -1.04 | 1.03 | -1.07 | 1.02 | 1.16 | -1.05 |
| hydrolase activity, acting on acid carbon-carbon bonds | 1 | 2.81 | -1.06 | -1.21 | -1.09 | 1.58 | 2.01 | -1.24 |
| hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances | 1 | 2.81 | -1.06 | -1.21 | -1.09 | 1.58 | 2.01 | -1.24 |
| hydrolase activity, acting on acid sulfur-nitrogen bonds | 1 | -1.48 | -1.35 | -1.19 | -1.31 | -1.2 | -1.3 | 1.23 |
| lyase activity | 125 | 1.07 | 1.01 | -1.04 | -1.02 | 1.05 | 1.11 | 1.03 |
| carbon-carbon lyase activity | 32 | 1.06 | -1.12 | -1.24 | 1.0 | -1.0 | 1.08 | -1.03 |
| carboxy-lyase activity | 25 | 1.06 | -1.19 | -1.28 | -1.05 | -1.03 | 1.05 | -1.03 |
| aldehyde-lyase activity | 4 | -1.25 | 1.0 | -1.2 | 1.35 | 1.18 | -1.18 | -1.45 |
| oxo-acid-lyase activity | 1 | -1.23 | 1.82 | 1.48 | 1.33 | 1.52 | 1.63 | 1.08 |
| carbon-oxygen lyase activity | 52 | 1.11 | 1.04 | 1.1 | -1.06 | 1.02 | 1.15 | 1.05 |
| hydro-lyase activity | 45 | 1.04 | 1.04 | 1.1 | -1.08 | -1.01 | 1.14 | 1.04 |
| carbon-oxygen lyase activity, acting on polysaccharides | 1 | 1.35 | 1.02 | 1.15 | 1.05 | 1.09 | 1.1 | 1.03 |
| carbon-oxygen lyase activity, acting on phosphates | 1 | 2.12 | 1.31 | 1.32 | 1.23 | 1.73 | 1.01 | 2.0 |
| carbon-nitrogen lyase activity | 5 | -2.25 | -1.05 | -1.88 | 1.19 | 2.05 | -1.63 | -1.2 |
| ammonia-lyase activity | 1 | -3.81 | -2.74 | -2.14 | -1.15 | 11.07 | -1.69 | -3.46 |
| amidine-lyase activity | 2 | -3.73 | 1.28 | -2.46 | 1.25 | 1.21 | -1.91 | 1.03 |
| amine-lyase activity | 2 | -1.04 | 1.14 | -1.34 | 1.32 | 1.5 | -1.36 | 1.15 |
| strictosidine synthase activity | 2 | -1.04 | 1.14 | -1.34 | 1.32 | 1.5 | -1.36 | 1.15 |
| carbon-sulfur lyase activity | 6 | 1.18 | 1.18 | 1.05 | 1.05 | 1.02 | 1.08 | -1.01 |
| 1-aminocyclopropane-1-carboxylate synthase activity | 2 | 1.14 | 1.17 | -1.17 | -1.01 | 1.02 | 1.07 | -1.0 |
| carbon-halide lyase activity | 1 | -1.05 | 1.06 | 1.25 | -1.2 | 1.21 | -1.46 | 1.19 |
| phosphorus-oxygen lyase activity | 28 | 1.14 | 1.04 | -1.01 | -1.01 | 1.03 | 1.2 | 1.09 |
| isomerase activity | 93 | 1.39 | 1.07 | 1.04 | 1.1 | 1.15 | 1.21 | 1.08 |
| racemase and epimerase activity | 9 | 1.08 | 1.08 | -1.25 | -1.0 | 1.14 | -1.19 | 1.12 |
| racemase and epimerase activity, acting on carbohydrates and derivatives | 8 | -1.09 | 1.13 | -1.29 | -1.06 | 1.05 | -1.3 | 1.17 |
| cis-trans isomerase activity | 29 | 1.39 | 1.03 | 1.12 | 1.15 | 1.24 | 1.15 | 1.02 |
| intramolecular oxidoreductase activity | 26 | 1.67 | 1.02 | 1.03 | 1.12 | 1.09 | 1.42 | 1.03 |
| intramolecular oxidoreductase activity, interconverting aldoses and ketoses | 5 | 2.55 | 1.17 | -1.07 | 1.0 | 1.07 | 1.9 | 1.31 |
| intramolecular oxidoreductase activity, interconverting keto- and enol-groups | 11 | 1.16 | 1.01 | 1.21 | 1.13 | 1.12 | 1.18 | 1.09 |
| intramolecular oxidoreductase activity, transposing C=C bonds | 10 | 2.03 | -1.04 | -1.1 | 1.17 | 1.08 | 1.51 | -1.15 |
| intramolecular oxidoreductase activity, transposing S-S bonds | 10 | 1.1 | -1.01 | 1.22 | 1.11 | 1.12 | 1.08 | 1.12 |
| intramolecular transferase activity | 19 | 1.31 | 1.16 | 1.03 | 1.05 | 1.07 | 1.29 | 1.12 |
| intramolecular transferase activity, phosphotransferases | 8 | 1.0 | 1.04 | 1.02 | 1.02 | 1.07 | -1.12 | 1.12 |
| intramolecular lyase activity | 1 | 1.86 | -1.46 | -1.12 | -1.51 | 1.13 | 1.83 | 1.71 |
| ligase activity | 206 | 1.04 | 1.13 | 1.16 | 1.09 | 1.14 | 1.1 | 1.07 |
| ligase activity, forming carbon-oxygen bonds | 40 | -1.07 | 1.26 | 1.26 | 1.14 | 1.18 | 1.05 | 1.09 |
| ligase activity, forming aminoacyl-tRNA and related compounds | 40 | -1.07 | 1.26 | 1.26 | 1.14 | 1.18 | 1.05 | 1.09 |
| ligase activity, forming carbon-sulfur bonds | 20 | -1.5 | -1.17 | -1.49 | -1.12 | -1.4 | -1.6 | -1.77 |
| acid-thiol ligase activity | 13 | -2.56 | -1.1 | -1.57 | -1.26 | -1.84 | -1.46 | -2.12 |
| ligase activity, forming carbon-nitrogen bonds | 138 | 1.14 | 1.14 | 1.23 | 1.1 | 1.21 | 1.21 | 1.16 |
| acid-ammonia (or amide) ligase activity | 2 | 1.91 | -1.99 | 2.01 | -1.44 | 1.52 | -1.73 | -1.25 |
| acid-amino acid ligase activity | 111 | 1.11 | 1.16 | 1.34 | 1.13 | 1.26 | 1.19 | 1.18 |
| cyclo-ligase activity | 3 | 1.09 | 1.06 | 1.12 | 1.21 | 1.12 | 1.24 | -1.09 |
| carbon-nitrogen ligase activity, with glutamine as amido-N-donor | 13 | 1.37 | 1.08 | -1.29 | -1.04 | -1.04 | 1.51 | 1.21 |
| ligase activity, forming carbon-carbon bonds | 4 | 2.44 | 1.26 | -1.08 | -1.11 | 1.01 | 1.91 | 1.62 |
| ligase activity, forming phosphoric ester bonds | 5 | 1.2 | 1.22 | 1.08 | 1.19 | 1.21 | 1.0 | 1.01 |
| endodeoxyribonuclease activity, producing 5'-phosphomonoesters | 2 | 2.29 | 1.05 | 1.61 | -1.06 | 1.36 | 1.86 | 1.38 |
| endodeoxyribonuclease activity, producing 3'-phosphomonoesters | 2 | 1.96 | -1.84 | -1.77 | -1.88 | -1.43 | -1.2 | 2.09 |
| endoribonuclease activity, producing 5'-phosphomonoesters | 12 | 1.14 | 1.09 | 1.3 | 1.19 | 1.29 | 1.24 | 1.0 |
| endoribonuclease activity, producing 3'-phosphomonoesters | 2 | 1.91 | 1.08 | 1.3 | 1.12 | 1.28 | 1.27 | 1.22 |
| endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters | 14 | 1.26 | 1.08 | 1.34 | 1.15 | 1.3 | 1.31 | 1.05 |
| endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 3'-phosphomonoesters | 4 | 1.94 | -1.3 | -1.17 | -1.3 | -1.06 | 1.03 | 1.6 |
| exodeoxyribonuclease activity, producing 5'-phosphomonoesters | 7 | -1.09 | 1.11 | 1.04 | 1.05 | 1.14 | -1.05 | -1.01 |
| exoribonuclease activity, producing 5'-phosphomonoesters | 18 | 1.38 | 1.17 | 1.2 | 1.26 | 1.28 | 1.2 | 1.08 |
| oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor | 1 | 1.11 | 1.03 | -1.05 | 1.12 | 4.21 | 1.26 | -1.02 |
| oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor | 1 | 1.07 | 1.03 | -1.07 | 1.05 | -1.1 | -1.02 | -1.18 |
| oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor | 5 | -1.02 | -1.05 | 1.01 | 1.08 | 1.13 | 1.19 | -1.09 |
| G-protein coupled acetylcholine receptor activity | 2 | 1.0 | 1.08 | 1.1 | 1.16 | 1.17 | 1.06 | 1.05 |
| SAP kinase activity | 5 | 1.03 | 1.11 | 1.44 | 1.2 | 1.45 | -1.1 | -1.04 |
| GABA receptor activity | 7 | 1.0 | 1.05 | -1.0 | -1.01 | -1.08 | -1.05 | -1.12 |
| ligand-dependent nuclear receptor binding | 5 | 1.26 | 1.45 | 1.61 | 1.27 | 1.41 | 1.59 | 1.27 |
| thyroid hormone receptor binding | 1 | -1.32 | 1.3 | -1.05 | 1.3 | 1.04 | 2.32 | -1.33 |
| protein sumoylation | 3 | 1.29 | 1.14 | 1.26 | 1.29 | 1.5 | 1.22 | -1.02 |
| SUMO-specific protease activity | 1 | 1.48 | 1.3 | 2.69 | 1.46 | 2.61 | 3.81 | 1.95 |
| extracellular-glycine-gated ion channel activity | 7 | -1.5 | 1.06 | -1.1 | -1.52 | -1.91 | -1.85 | 1.36 |
| extracellular-glycine-gated chloride channel activity | 4 | -1.54 | 1.26 | 1.47 | -1.43 | -1.72 | -1.63 | -1.04 |
| glycine-gated chloride channel complex | 4 | -1.54 | 1.26 | 1.47 | -1.43 | -1.72 | -1.63 | -1.04 |
| short-branched-chain-acyl-CoA dehydrogenase activity | 2 | -1.12 | 1.32 | -1.16 | 1.05 | 1.12 | -1.93 | 1.2 |
| kinesin II complex | 1 | 1.07 | 1.09 | 1.33 | 1.24 | 1.41 | 1.98 | -1.06 |
| natriuretic peptide receptor activity | 1 | -1.36 | 1.26 | 1.17 | 1.07 | -1.37 | -1.35 | 1.16 |
| insulin-like growth factor binding protein complex | 2 | -1.01 | -1.08 | -1.05 | -1.02 | 1.09 | -1.07 | -1.03 |
| flavin-linked sulfhydryl oxidase activity | 5 | -1.04 | 1.15 | 1.04 | 1.11 | 1.03 | 1.33 | -1.08 |
| thiol oxidase activity | 5 | -1.04 | 1.15 | 1.04 | 1.11 | 1.03 | 1.33 | -1.08 |
| poly(A)+ mRNA export from nucleus | 6 | 1.05 | 1.1 | 1.12 | 1.19 | 1.19 | -1.03 | -1.02 |
| lipoate-protein ligase activity | 2 | 1.65 | 1.07 | 1.42 | 1.08 | 1.12 | 2.67 | -1.13 |
| lipoate synthase activity | 1 | 1.58 | 1.18 | 1.95 | 1.1 | 1.45 | 3.43 | -1.02 |
| cell wall macromolecule catabolic process | 9 | -1.18 | -1.16 | -1.42 | -1.31 | -1.28 | -2.35 | -1.08 |
| antibiotic metabolic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| antibiotic biosynthetic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| activin-activated receptor activity | 2 | -1.14 | 1.57 | 1.93 | 1.23 | 1.67 | 1.43 | 1.4 |
| protein-heme linkage | 1 | 1.66 | 1.03 | 1.71 | 1.06 | 1.0 | 2.28 | -1.41 |
| cytochrome complex assembly | 3 | 1.45 | 1.07 | 1.3 | 1.03 | 1.04 | 1.58 | -1.23 |
| 3'-tyrosyl-DNA phosphodiesterase activity | 1 | -1.61 | -1.07 | -1.0 | -1.17 | -1.28 | -1.58 | -1.34 |
| protein-tetrapyrrole linkage | 1 | 1.66 | 1.03 | 1.71 | 1.06 | 1.0 | 2.28 | -1.41 |
| regulation of transforming growth factor beta receptor signaling pathway | 9 | 1.43 | 1.09 | 1.33 | -1.08 | 1.27 | 1.44 | 1.24 |
| Ras GTPase binding | 26 | 1.1 | 1.19 | 1.47 | 1.12 | 1.32 | 1.31 | 1.35 |
| MAP kinase tyrosine/serine/threonine phosphatase activity | 3 | 1.18 | 1.17 | 1.3 | 1.08 | 1.35 | 1.14 | 1.33 |
| myosin phosphatase activity | 3 | 1.52 | 1.01 | 1.94 | -1.04 | 1.33 | 1.77 | 1.57 |
| myosin phosphatase regulator activity | 1 | -3.34 | 1.3 | 1.86 | 1.22 | 1.27 | -2.0 | 1.07 |
| myosin binding | 23 | -1.09 | 1.05 | 1.41 | 1.02 | 1.21 | 1.04 | 1.08 |
| myosin phosphatase complex | 1 | 5.19 | -1.26 | 2.54 | -1.39 | 1.19 | 2.56 | 1.75 |
| TBP-class protein binding | 3 | 1.62 | 1.22 | 1.38 | 1.2 | 1.3 | 1.46 | 1.18 |
| protein stabilization | 15 | 1.39 | 1.17 | 1.1 | 1.12 | 1.19 | 1.04 | 1.03 |
| potassium:amino acid symporter activity | 5 | -1.79 | -1.85 | -2.57 | -1.19 | 1.65 | 1.54 | 1.09 |
| Rap guanyl-nucleotide exchange factor activity | 1 | -1.08 | 1.15 | 1.11 | 1.03 | 1.11 | -1.08 | 1.02 |
| protein import | 66 | 1.08 | 1.15 | 1.27 | 1.17 | 1.26 | 1.18 | 1.02 |
| positive regulation of hormone secretion | 1 | 1.12 | 1.15 | 1.24 | 1.0 | 1.0 | 1.1 | -1.06 |
| peptide hormone binding | 2 | 1.14 | 1.03 | 1.06 | 1.02 | -1.01 | -1.0 | -1.03 |
| Rho GTPase binding | 7 | 1.11 | 1.18 | 1.37 | 1.08 | 1.39 | 1.46 | 1.35 |
| GTP-Rho binding | 1 | -1.55 | 1.88 | -1.17 | -1.09 | -1.11 | -1.68 | 1.84 |
| D-erythro-sphingosine kinase activity | 3 | 2.17 | 1.08 | -1.12 | -1.03 | -1.02 | 1.22 | 1.39 |
| retinol dehydratase activity | 2 | 1.19 | -1.09 | -1.04 | -1.04 | 1.02 | 1.11 | -1.06 |
| transcriptional repressor complex | 14 | 1.34 | 1.2 | 1.42 | 1.15 | 1.41 | 1.35 | 1.31 |
| negative cofactor 2 complex | 2 | 1.34 | 1.17 | 1.22 | 1.42 | 1.42 | 1.02 | -1.0 |
| negative regulation of RNA polymerase II transcriptional preinitiation complex assembly | 3 | 1.71 | 1.26 | 1.41 | 1.3 | 1.44 | 1.38 | 1.2 |
| structural constituent of nuclear pore | 4 | 1.27 | 1.07 | 1.32 | 1.35 | 1.49 | 1.29 | 1.1 |
| 6-phosphogluconolactonase activity | 1 | 2.54 | 1.2 | -1.43 | 1.17 | 1.05 | -1.15 | 1.18 |
| FH1 domain binding | 2 | 1.0 | 1.2 | 1.28 | 1.34 | 1.19 | -1.05 | -1.08 |
| serine C-palmitoyltransferase complex | 1 | -1.92 | 1.11 | 2.34 | 1.27 | 1.64 | -1.44 | -1.26 |
| S-methyl-5-thioadenosine phosphorylase activity | 2 | -1.31 | 1.03 | -1.18 | 1.26 | 1.0 | -1.49 | -1.34 |
| respiratory chain complex III assembly | 1 | 1.04 | 1.04 | 1.01 | -1.04 | -1.1 | 1.13 | -1.12 |
| fatty acid amide hydrolase activity | 4 | 1.42 | 1.22 | -1.76 | -1.03 | -1.09 | 2.22 | 1.2 |
| single-strand selective uracil DNA N-glycosylase activity | 1 | 1.26 | 1.08 | -1.14 | 1.16 | 1.06 | -1.01 | -1.01 |
| tyrosine-ester sulfotransferase activity | 1 | -11.7 | 1.38 | -1.27 | 1.16 | 1.17 | -8.03 | 1.02 |
| U6 snRNA binding | 2 | 1.33 | 1.17 | 1.11 | 1.38 | 1.29 | 1.09 | -1.1 |
| intracellular cyclic nucleotide activated cation channel complex | 2 | -1.19 | -1.0 | 1.11 | -1.1 | 1.07 | 1.05 | -1.33 |
| syntaxin-1 binding | 1 | -1.34 | 1.17 | 1.4 | 1.35 | 1.51 | 1.53 | 1.84 |
| purine nucleotide binding | 813 | 1.08 | 1.1 | 1.19 | 1.09 | 1.22 | 1.17 | 1.09 |
| oxidative phosphorylation uncoupler activity | 5 | -1.29 | -1.01 | -1.03 | -1.02 | 1.03 | -1.07 | -1.18 |
| heat shock protein binding | 40 | 1.19 | 1.15 | 1.18 | 1.12 | 1.14 | 1.12 | 1.13 |
| sodium channel regulator activity | 1 | -1.11 | -1.01 | 1.04 | -1.05 | -1.05 | -1.14 | -1.0 |
| delta1-pyrroline-5-carboxylate synthetase activity | 1 | -29.94 | 1.93 | -2.31 | -1.05 | -1.13 | -7.63 | 1.24 |
| 3-methyl-2-oxobutanoate dehydrogenase (lipoamide) complex | 2 | 1.67 | 1.29 | 1.69 | -1.02 | 1.34 | 1.99 | 1.11 |
| mitochondrial processing peptidase complex | 1 | -1.11 | 1.21 | 1.34 | -1.03 | -1.05 | 2.16 | -1.38 |
| glycolipid transporter activity | 1 | -2.99 | 1.35 | -1.24 | 1.41 | -1.1 | -2.55 | -1.06 |
| meprin A complex | 4 | -1.05 | -1.07 | -1.08 | 1.07 | 1.0 | -1.08 | -1.08 |
| AU-rich element binding | 1 | -1.76 | 1.11 | 1.26 | 1.23 | 1.05 | -1.22 | -1.69 |
| heparan sulfate 6-O-sulfotransferase activity | 1 | 1.37 | -1.51 | 7.55 | 1.04 | 1.53 | 1.73 | 1.66 |
| sulfonylurea receptor binding | 1 | 2.98 | -1.49 | 1.51 | -1.4 | 1.16 | 14.12 | 1.01 |
| very-long-chain-acyl-CoA dehydrogenase activity | 1 | 1.8 | 1.71 | -1.79 | -1.14 | 1.11 | -1.09 | 1.11 |
| aminoacyl-tRNA synthetase multienzyme complex | 3 | 1.11 | 1.2 | 1.09 | 1.15 | 1.05 | -1.07 | 1.06 |
| anion exchanger adaptor activity | 1 | -1.15 | 1.11 | 1.03 | 1.13 | 1.14 | 1.45 | 1.27 |
| glutamate-cysteine ligase complex | 3 | -1.79 | 1.47 | -1.22 | 1.17 | -1.04 | -1.53 | 1.26 |
| Rab guanyl-nucleotide exchange factor activity | 2 | -1.44 | 1.17 | 1.82 | 1.09 | 1.3 | 1.78 | 1.08 |
| dihydropyrimidine dehydrogenase (NADP+) activity | 1 | -2.4 | 1.52 | -1.09 | 1.53 | -1.18 | -2.54 | -1.54 |
| peptidase inhibitor activity | 76 | 1.39 | -1.53 | -1.49 | -1.55 | -1.32 | 1.49 | 1.05 |
| single-stranded DNA-dependent ATP-dependent DNA helicase activity | 3 | 1.08 | 1.14 | 1.41 | 1.15 | 1.37 | -1.0 | 1.15 |
| single-stranded DNA-dependent ATP-dependent DNA helicase complex | 1 | -1.41 | 1.08 | 1.3 | 1.52 | 1.33 | -1.66 | -1.08 |
| lipoyltransferase activity | 1 | 1.33 | -1.19 | 1.85 | 1.0 | 1.1 | 2.24 | -1.59 |
| Golgi transport complex | 2 | 1.23 | 1.33 | 1.33 | 1.17 | 1.27 | 1.38 | 1.89 |
| protein N-acetylglucosaminyltransferase complex | 2 | 1.22 | 1.13 | 1.2 | 1.2 | 1.16 | 1.19 | 1.37 |
| Ral GTPase activator activity | 1 | 1.09 | 1.26 | 1.08 | 1.12 | 1.12 | 1.3 | 1.36 |
| SH3 domain binding | 8 | 1.15 | 1.04 | 1.33 | 1.04 | 1.19 | 1.55 | 1.28 |
| deoxycytidyl transferase activity | 1 | 1.75 | -1.08 | 2.07 | 1.17 | 1.37 | 2.15 | 1.01 |
| nucleologenesis | 1 | 1.32 | 1.42 | 1.69 | 1.17 | 1.76 | 1.08 | 1.69 |
| cholesterol transporter activity | 1 | -1.06 | 1.09 | -1.0 | 1.4 | 1.63 | -1.48 | -1.12 |
| phospholipid scramblase activity | 2 | 2.57 | -1.14 | 1.03 | -1.07 | 2.13 | 1.16 | 1.16 |
| triglyceride binding | 1 | 4.36 | -1.08 | -1.33 | -1.1 | 1.06 | 2.49 | 1.49 |
| uridine-rich cytoplasmic polyadenylylation element binding | 1 | 1.17 | -1.07 | -1.05 | 1.02 | -1.05 | -1.03 | -1.08 |
| cyclic nucleotide-dependent guanyl-nucleotide exchange factor activity | 2 | -1.1 | 1.13 | 1.09 | -1.05 | -1.04 | -1.02 | -1.01 |
| mitochondrial electron transfer flavoprotein complex | 2 | 1.33 | 1.29 | -1.39 | 1.04 | 1.01 | -1.08 | -1.3 |
| fibroblast growth factor binding | 1 | 3.07 | 1.39 | 2.07 | 1.3 | 2.02 | 1.75 | 1.45 |
| enzyme binding | 116 | 1.3 | 1.11 | 1.41 | 1.1 | 1.33 | 1.31 | 1.26 |
| NAD-dependent histone deacetylase activity | 2 | 1.85 | 1.16 | 1.29 | 1.09 | 1.23 | 1.67 | 1.46 |
| Rab GTPase binding | 6 | 1.23 | 1.16 | 1.16 | 1.13 | 1.22 | 1.16 | 1.33 |
| response to arsenic-containing substance | 8 | 1.07 | 1.17 | 1.52 | 1.13 | 1.46 | 1.14 | 1.14 |
| insecticide metabolic process | 10 | 1.1 | -1.02 | -1.79 | -1.06 | -1.28 | -1.46 | 1.06 |
| drug metabolic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| stem cell division | 64 | -1.0 | 1.13 | 1.32 | 1.11 | 1.34 | 1.07 | 1.11 |
| N-methyl-D-aspartate selective glutamate receptor complex | 3 | -3.51 | 1.06 | 1.06 | -1.04 | 1.07 | -1.9 | 1.09 |
| Wnt-protein binding | 11 | 1.57 | -1.01 | 1.22 | -1.02 | 1.31 | -1.0 | 1.28 |
| negative regulation of translation | 26 | 1.35 | 1.03 | 1.33 | 1.02 | 1.32 | 1.38 | 1.31 |
| tRNA dihydrouridine synthase activity | 4 | 1.57 | 1.16 | 1.09 | 1.25 | 1.12 | 1.12 | -1.03 |
| DEAD/H-box RNA helicase binding | 2 | 1.24 | 1.45 | 1.49 | 1.18 | 1.41 | 1.54 | 1.43 |
| sodium:dicarboxylate symporter activity | 3 | 1.33 | 1.26 | -3.33 | -3.11 | -2.74 | -1.43 | 1.86 |
| semaphorin receptor activity | 2 | 1.69 | 1.12 | 2.14 | -1.0 | 1.95 | 2.34 | 1.53 |
| regulation of exocytosis | 9 | -1.22 | 1.13 | 1.23 | 1.07 | 1.14 | 1.53 | 1.05 |
| regulation of calcium ion-dependent exocytosis | 1 | -1.33 | -1.11 | -1.13 | -1.17 | -1.09 | 12.66 | 1.12 |
| pantetheine hydrolase activity | 3 | -3.81 | -1.17 | -3.75 | 2.31 | -1.17 | -3.72 | -1.67 |
| Ral GTPase binding | 3 | 1.22 | 1.31 | 1.26 | 1.22 | 1.29 | 1.46 | 1.42 |
| aryl hydrocarbon receptor binding | 1 | 1.71 | 1.13 | 1.25 | 1.13 | 1.02 | 1.11 | -1.02 |
| vinculin binding | 1 | 2.93 | 1.13 | 1.23 | -1.06 | 1.04 | 3.7 | 1.38 |
| 5-oxoprolinase (ATP-hydrolyzing) activity | 1 | -1.27 | -1.09 | -1.84 | 1.84 | -1.17 | -2.01 | -1.59 |
| CDP-alcohol phosphatidyltransferase activity | 6 | -1.0 | 1.26 | 1.69 | 1.08 | 1.28 | -1.12 | 1.22 |
| KU70 binding | 1 | -1.09 | -1.0 | 1.57 | 1.36 | 1.26 | 1.16 | -2.0 |
| serine hydrolase activity | 290 | -2.22 | -1.39 | -1.61 | -1.12 | -1.45 | -2.28 | -1.59 |
| cysteine dioxygenase activity | 1 | 1.24 | 1.05 | 1.09 | -1.01 | -1.04 | 1.21 | 1.11 |
| glycine N-methyltransferase activity | 1 | 1.02 | -2.19 | -1.44 | -1.75 | -1.84 | 4.35 | -2.08 |
| phosphatidylinositol N-acetylglucosaminyltransferase activity | 4 | 1.26 | 1.36 | 1.66 | 1.37 | 1.48 | -1.0 | 1.01 |
| glucosidase II complex | 4 | -1.98 | 1.19 | -1.8 | 1.24 | 1.01 | 1.0 | -1.01 |
| peptidyl-diphthamide metabolic process | 3 | 1.96 | 1.14 | 1.33 | 1.22 | 1.29 | 1.28 | -1.07 |
| peptidyl-diphthamide biosynthetic process from peptidyl-histidine | 3 | 1.96 | 1.14 | 1.33 | 1.22 | 1.29 | 1.28 | -1.07 |
| peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase | 2 | 2.71 | -1.35 | -1.37 | -1.7 | -1.26 | -1.18 | 2.94 |
| peptidyl-glutamic acid carboxylation | 1 | 1.13 | 1.29 | 1.08 | 1.08 | -1.09 | -1.02 | 1.13 |
| N-terminal peptidyl-methionine acetylation | 1 | -1.5 | 1.47 | 2.65 | 1.15 | 1.31 | -1.14 | -1.06 |
| N-terminal peptidyl-L-cysteine N-palmitoylation | 1 | 1.07 | 1.32 | 1.75 | 1.48 | 1.85 | -1.07 | -1.03 |
| peptidyl-lysine methylation | 4 | 1.32 | 1.26 | 1.73 | 1.16 | 1.81 | 1.88 | 1.52 |
| peptidyl-lysine trimethylation | 3 | 1.35 | 1.23 | 1.68 | 1.16 | 1.73 | 1.61 | 1.45 |
| histone-lysine N-methyltransferase activity | 14 | 1.29 | 1.27 | 1.5 | 1.07 | 1.4 | 1.43 | 1.38 |
| peptidyl-lysine monomethylation | 1 | 1.22 | 1.34 | 1.86 | 1.14 | 2.08 | 2.98 | 1.75 |
| peptidyl-lysine dimethylation | 1 | 1.41 | 1.59 | 1.77 | 1.23 | 1.83 | 1.65 | 1.69 |
| cytochrome c-heme linkage | 1 | 1.66 | 1.03 | 1.71 | 1.06 | 1.0 | 2.28 | -1.41 |
| protein-cofactor linkage | 1 | -1.1 | 1.02 | -1.19 | -1.23 | -1.41 | 1.01 | -1.14 |
| protein polyglycylation | 2 | -1.15 | -1.05 | -1.18 | -1.0 | -1.15 | -1.19 | -1.18 |
| peptidyl-serine phosphorylation | 3 | 1.28 | 1.07 | 1.17 | 1.15 | 1.14 | 1.08 | -1.08 |
| peptidyl-histidine phosphorylation | 1 | 1.67 | -1.54 | 1.2 | -1.13 | 7.02 | 3.44 | 1.09 |
| peptidyl-threonine phosphorylation | 1 | 1.19 | -1.04 | 1.15 | 1.11 | 1.6 | 1.03 | 1.09 |
| peptidyl-tyrosine phosphorylation | 5 | 1.6 | 1.02 | 1.5 | 1.02 | 1.17 | 1.5 | 1.38 |
| heterocycle biosynthetic process | 146 | 1.22 | 1.04 | 1.02 | -1.01 | 1.0 | 1.25 | 1.03 |
| peptide cross-linking | 3 | 3.33 | -1.61 | -1.45 | -1.67 | -1.2 | 1.72 | 1.67 |
| peptidyl-pyrromethane cofactor linkage | 1 | -1.1 | 1.02 | -1.19 | -1.23 | -1.41 | 1.01 | -1.14 |
| C-terminal protein-tyrosinylation | 1 | 1.09 | -1.0 | -1.03 | 1.18 | -1.04 | 1.05 | -1.03 |
| peptidyl-amino acid modification | 62 | 1.26 | 1.09 | 1.2 | 1.06 | 1.19 | 1.24 | 1.17 |
| peptidyl-arginine modification | 3 | 1.19 | 1.12 | 1.45 | 1.13 | 1.36 | 1.2 | 1.09 |
| peptidyl-asparagine modification | 3 | -1.19 | 1.11 | 1.12 | 1.13 | 1.21 | 1.27 | 1.61 |
| peptidyl-cysteine modification | 2 | -1.01 | 1.16 | 1.21 | 1.1 | 1.15 | -1.03 | -1.08 |
| peptidyl-glutamine modification | 2 | 2.71 | -1.35 | -1.37 | -1.7 | -1.26 | -1.18 | 2.94 |
| peptidyl-glutamic acid modification | 1 | 1.13 | 1.29 | 1.08 | 1.08 | -1.09 | -1.02 | 1.13 |
| peptidyl-histidine modification | 4 | 1.89 | -1.01 | 1.3 | 1.12 | 1.97 | 1.64 | -1.03 |
| peptidyl-lysine modification | 20 | 1.35 | 1.22 | 1.43 | 1.16 | 1.42 | 1.47 | 1.29 |
| peptidyl-methionine modification | 2 | 1.17 | 1.23 | 1.52 | -1.02 | 1.0 | -1.03 | -1.15 |
| peptidyl-proline modification | 15 | -1.0 | 1.02 | -1.1 | -1.04 | -1.12 | 1.01 | -1.04 |
| peptidyl-threonine modification | 1 | 1.19 | -1.04 | 1.15 | 1.11 | 1.6 | 1.03 | 1.09 |
| peptidyl-tyrosine modification | 6 | 1.5 | 1.01 | 1.39 | 1.05 | 1.14 | 1.41 | 1.3 |
| protein carboxylation | 1 | 1.13 | 1.29 | 1.08 | 1.08 | -1.09 | -1.02 | 1.13 |
| protein phosphopantetheinylation | 1 | 1.84 | 1.09 | 2.09 | 1.3 | 2.04 | 2.34 | 1.11 |
| peptidyl-arginine methylation | 3 | 1.19 | 1.12 | 1.45 | 1.13 | 1.36 | 1.2 | 1.09 |
| biotin-protein ligase activity | 1 | 1.43 | 1.46 | 1.44 | 1.24 | 1.28 | 1.18 | 1.38 |
| protein N-linked glycosylation via asparagine | 3 | -1.19 | 1.11 | 1.12 | 1.13 | 1.21 | 1.27 | 1.61 |
| protein tyrosinylation | 1 | 1.09 | -1.0 | -1.03 | 1.18 | -1.04 | 1.05 | -1.03 |
| protein palmitoylation | 22 | -1.11 | 1.1 | 1.13 | 1.12 | 1.09 | -1.16 | -1.11 |
| protein myristoylation | 1 | -1.49 | 1.29 | 1.71 | 1.24 | 1.7 | 1.05 | 1.0 |
| glycoprotein 3-alpha-L-fucosyltransferase activity | 1 | -1.29 | 1.13 | -1.16 | 1.54 | 2.78 | 1.39 | 1.18 |
| internal peptidyl-lysine acetylation | 14 | 1.4 | 1.24 | 1.42 | 1.18 | 1.37 | 1.44 | 1.29 |
| peptidyl-lysine acetylation | 14 | 1.4 | 1.24 | 1.42 | 1.18 | 1.37 | 1.44 | 1.29 |
| peptidyl-proline hydroxylation to 4-hydroxy-L-proline | 15 | -1.0 | 1.02 | -1.1 | -1.04 | -1.12 | 1.01 | -1.04 |
| C-terminal protein amino acid modification | 2 | -1.4 | 1.08 | 1.03 | 1.29 | 1.32 | -1.28 | -1.35 |
| translation release factor complex | 1 | 1.2 | 1.24 | 1.0 | 1.1 | -1.06 | 1.25 | 1.2 |
| prothoracicotrophic hormone activity | 1 | -1.06 | -1.47 | -1.01 | -1.23 | -1.12 | 1.54 | -1.14 |
| acetoacetyl-CoA reductase activity | 1 | 1.03 | -1.04 | 1.2 | -1.01 | 1.0 | 2.11 | -1.07 |
| alkane 1-monooxygenase activity | 1 | 1.6 | 1.35 | -1.72 | -1.6 | -1.17 | -1.33 | 1.5 |
| S-formylglutathione hydrolase activity | 1 | 1.41 | 1.06 | -1.13 | 1.33 | 1.12 | -1.09 | -1.51 |
| alkyl sulfatase activity | 1 | 2.4 | 1.23 | 1.72 | -1.4 | 3.44 | 4.52 | 1.64 |
| DDT-dehydrochlorinase activity | 1 | -1.05 | 1.06 | 1.25 | -1.2 | 1.21 | -1.46 | 1.19 |
| 1-aminocyclopropane-1-carboxylate metabolic process | 2 | 1.14 | 1.17 | -1.17 | -1.01 | 1.02 | 1.07 | -1.0 |
| organic ether metabolic process | 26 | 1.41 | -1.13 | -1.09 | -1.07 | 1.17 | -1.17 | 1.01 |
| phenol-containing compound metabolic process | 9 | -1.02 | -1.18 | -1.26 | -1.16 | -1.28 | -1.1 | 1.52 |
| molting cycle, protein-based cuticle | 55 | 1.16 | -1.03 | 1.01 | -1.09 | -1.04 | 1.19 | 1.17 |
| ecdysis, chitin-based cuticle | 7 | 1.02 | 1.08 | 1.04 | -1.01 | -1.03 | 1.01 | 1.06 |
| oviposition | 12 | 1.07 | 1.1 | -1.02 | -1.01 | -1.06 | -1.03 | -1.02 |
| germ-line sex determination | 5 | 1.56 | 1.08 | 1.18 | 1.1 | 1.06 | 1.51 | 1.13 |
| somatic sex determination | 17 | 1.27 | 1.15 | 1.28 | 1.11 | 1.21 | 1.09 | 1.12 |
| guanyl nucleotide binding | 161 | 1.17 | 1.08 | 1.28 | 1.07 | 1.23 | 1.1 | 1.07 |
| GDP binding | 3 | 2.03 | -1.04 | 1.6 | 1.01 | 1.54 | 1.95 | 1.24 |
| molybdopterin synthase complex | 1 | 2.51 | 1.28 | 2.88 | 1.18 | 3.18 | 1.65 | 1.8 |
| farnesoic acid O-methyltransferase activity | 2 | 3.79 | -5.16 | -3.85 | -4.65 | -1.93 | 1.64 | 2.51 |
| virion | 7 | -1.09 | -1.36 | -1.17 | -1.1 | -1.08 | 1.28 | -1.28 |
| viral capsid | 6 | -1.3 | -1.35 | -1.31 | -1.0 | -1.02 | -1.17 | -1.33 |
| viral envelope | 1 | 2.63 | -1.41 | 1.69 | -1.89 | -1.48 | 14.61 | -1.02 |
| viral envelope fusion with host membrane | 1 | 2.63 | -1.41 | 1.69 | -1.89 | -1.48 | 14.61 | -1.02 |
| virus-host interaction | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| viral infectious cycle | 3 | -1.12 | 1.07 | 1.27 | -1.16 | -1.11 | 1.69 | 1.21 |
| initiation of viral infection | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| mitochondrial rRNA export from mitochondrion | 1 | 3.27 | 1.48 | 4.47 | -1.01 | 2.24 | 5.65 | 2.22 |
| mitochondrial RNA localization | 1 | 3.27 | 1.48 | 4.47 | -1.01 | 2.24 | 5.65 | 2.22 |
| pole plasm mRNA localization | 37 | 1.02 | 1.19 | 1.31 | 1.15 | 1.26 | 1.21 | 1.17 |
| reproductive behavior | 89 | 1.01 | -1.08 | -1.02 | -1.04 | -1.07 | 1.01 | 1.09 |
| female germ-line sex determination | 4 | 1.82 | 1.02 | 1.1 | 1.07 | 1.07 | 1.73 | 1.13 |
| female somatic sex determination | 5 | 1.87 | 1.25 | 1.34 | 1.23 | 1.35 | 1.16 | 1.18 |
| male somatic sex determination | 2 | 2.22 | 1.29 | 1.1 | 1.16 | 1.11 | -1.15 | 1.06 |
| myristoyltransferase activity | 1 | -1.49 | 1.29 | 1.71 | 1.24 | 1.7 | 1.05 | 1.0 |
| deoxynucleoside kinase activity | 1 | 1.02 | 1.09 | 1.32 | 1.32 | 1.3 | -1.29 | -1.13 |
| trans-2-enoyl-CoA reductase (NADPH) activity | 1 | 1.29 | -1.03 | 1.55 | 1.16 | 1.07 | 1.93 | -1.33 |
| 3-hydroxyacyl-[acyl-carrier-protein] dehydratase activity | 2 | 3.49 | -1.07 | -1.08 | -1.15 | 1.4 | 1.97 | 2.1 |
| histamine-gated chloride channel activity | 2 | -1.17 | 1.03 | -1.13 | 1.06 | -1.2 | -1.2 | -1.2 |
| histamine-gated chloride channel complex | 2 | -1.17 | 1.03 | -1.13 | 1.06 | -1.2 | -1.2 | -1.2 |
| nonribosomal peptide biosynthetic process | 1 | 1.36 | 1.09 | 1.02 | -1.03 | -1.16 | 1.04 | 1.01 |
| snRNA-activating protein complex | 2 | 1.7 | 1.14 | 1.37 | 1.1 | 1.36 | 1.18 | 1.22 |
| acyl-CoA N-acyltransferase activity | 1 | 1.2 | 1.15 | 1.75 | 1.03 | 1.4 | 1.08 | 1.36 |
| beta-1,4-mannosyltransferase activity | 2 | -2.16 | 1.25 | 1.27 | 1.12 | 1.95 | -1.56 | -1.08 |
| transmembrane receptor protein phosphatase activity | 6 | 1.57 | 1.05 | 1.23 | 1.03 | 1.35 | 2.22 | 1.25 |
| transmembrane receptor protein kinase activity | 26 | 1.41 | 1.03 | 1.2 | 1.02 | 1.21 | 1.47 | 1.43 |
| carbohydrate kinase activity | 14 | 1.04 | 1.03 | -1.21 | 1.04 | 1.14 | 1.34 | -1.15 |
| nucleotide kinase activity | 13 | 1.94 | -1.05 | 1.14 | -1.01 | 1.08 | 1.44 | -1.06 |
| amino acid kinase activity | 4 | -2.33 | 1.24 | -1.02 | 1.02 | -1.11 | -1.42 | -1.1 |
| carbohydrate phosphatase activity | 5 | 3.03 | -1.36 | 1.1 | -1.47 | -1.11 | 2.72 | 1.19 |
| nucleotide phosphatase activity | 3 | 1.37 | 1.12 | -1.02 | 1.28 | 1.23 | -1.07 | 1.06 |
| nucleobase-containing compound kinase activity | 24 | 1.52 | -1.0 | 1.19 | 1.05 | 1.13 | 1.31 | -1.03 |
| nucleoside kinase activity | 7 | 1.64 | -1.1 | 1.15 | -1.01 | 1.03 | 1.51 | 1.03 |
| kinase regulator activity | 45 | 1.15 | 1.13 | 1.2 | 1.14 | 1.29 | 1.05 | 1.1 |
| phosphatase regulator activity | 34 | 1.39 | -1.03 | 1.21 | 1.04 | 1.21 | 1.41 | 1.13 |
| kinase activator activity | 5 | 1.59 | 1.12 | 1.23 | 1.13 | 1.26 | 1.27 | 1.08 |
| kinase inhibitor activity | 6 | 1.32 | 1.13 | 1.13 | 1.05 | 1.2 | 1.27 | 1.35 |
| phosphatase activator activity | 3 | 1.39 | 1.03 | 1.11 | 1.1 | 1.16 | 1.44 | 1.03 |
| phosphatase inhibitor activity | 9 | 1.54 | -1.03 | 1.13 | 1.03 | 1.34 | 1.35 | 1.05 |
| deacetylase activity | 12 | 1.36 | 1.08 | 1.02 | 1.17 | 1.14 | 1.18 | 1.05 |
| regulation of lipid metabolic process | 9 | -2.45 | -1.13 | 1.14 | 1.12 | 1.3 | -2.52 | -1.85 |
| regulation of fatty acid metabolic process | 1 | 1.18 | 1.17 | 1.43 | 1.18 | 1.36 | -1.03 | 1.42 |
| regulation of steroid metabolic process | 1 | -1.55 | 1.2 | 1.21 | -1.19 | -1.12 | 1.05 | 1.03 |
| regulation of nucleobase-containing compound metabolic process | 796 | 1.13 | 1.09 | 1.18 | 1.07 | 1.17 | 1.17 | 1.12 |
| regulation of phosphate metabolic process | 80 | 1.29 | 1.13 | 1.33 | 1.09 | 1.29 | 1.35 | 1.2 |
| regulation of metabolic process | 1137 | 1.19 | 1.08 | 1.19 | 1.06 | 1.18 | 1.19 | 1.13 |
| transmission of nerve impulse | 223 | 1.08 | 1.07 | 1.2 | 1.03 | 1.19 | 1.22 | 1.13 |
| neuronal action potential propagation | 1 | -1.57 | 1.18 | 1.51 | 1.08 | 1.08 | -1.02 | 1.18 |
| regulation of action potential in neuron | 6 | 1.77 | -1.07 | 1.38 | -1.13 | 1.24 | 3.0 | 1.46 |
| sensory perception of pain | 2 | 2.05 | -1.17 | 1.22 | 1.13 | 1.44 | 2.13 | 1.2 |
| response to pheromone | 18 | 1.24 | -1.22 | -1.2 | -1.21 | -1.23 | 1.03 | -1.13 |
| cyclohydrolase activity | 4 | 2.83 | -1.76 | -1.94 | -1.6 | -1.8 | 1.77 | 1.28 |
| deaminase activity | 18 | -1.26 | 1.14 | -1.1 | -1.04 | -1.01 | 1.04 | -1.06 |
| mannose biosynthetic process | 1 | -1.31 | 1.22 | 1.33 | 1.17 | 1.22 | -1.88 | 1.41 |
| inositol catabolic process | 1 | 3.92 | -4.04 | -3.69 | -3.19 | -2.14 | 5.76 | -2.59 |
| hexose metabolic process | 62 | -1.54 | -1.05 | -1.7 | 1.0 | -1.08 | -1.5 | -1.54 |
| hexose biosynthetic process | 7 | -1.14 | -1.06 | -1.03 | -1.11 | 1.17 | 1.01 | -1.0 |
| hexose catabolic process | 33 | 1.52 | 1.06 | -1.18 | -1.03 | -1.06 | 1.37 | 1.2 |
| pentose metabolic process | 5 | 1.48 | 1.18 | -1.47 | 1.14 | -1.1 | -1.06 | 1.13 |
| phenol-containing compound catabolic process | 1 | -8.98 | 1.77 | -2.05 | -1.02 | -2.08 | -14.22 | 1.22 |
| cysteine biosynthetic process via cystathionine | 1 | 1.25 | 1.1 | 1.48 | -1.98 | -7.11 | -1.17 | 2.59 |
| cysteine biosynthetic process | 1 | 1.25 | 1.1 | 1.48 | -1.98 | -7.11 | -1.17 | 2.59 |
| nicotinate nucleotide biosynthetic process | 1 | 8.31 | 1.05 | -1.79 | -1.54 | -1.19 | 1.17 | 2.54 |
| nicotinate nucleotide salvage | 1 | 8.31 | 1.05 | -1.79 | -1.54 | -1.19 | 1.17 | 2.54 |
| nicotinamide nucleotide biosynthetic process | 3 | 1.9 | 1.5 | -1.09 | 1.01 | 1.09 | 1.23 | 1.86 |
| pyridine nucleotide metabolic process | 11 | 1.88 | 1.17 | -1.48 | -1.05 | 1.08 | 1.25 | 1.36 |
| pyridine nucleotide biosynthetic process | 3 | 1.9 | 1.5 | -1.09 | 1.01 | 1.09 | 1.23 | 1.86 |
| pyridine nucleotide salvage | 1 | 8.31 | 1.05 | -1.79 | -1.54 | -1.19 | 1.17 | 2.54 |
| fatty acid elongation, unsaturated fatty acid | 1 | -5.51 | -5.31 | -5.23 | -5.58 | -5.15 | -5.86 | 15.65 |
| leukotriene biosynthetic process | 1 | -1.74 | 1.11 | -2.01 | 1.51 | 1.27 | -1.87 | -1.55 |
| glycolipid catabolic process | 1 | -1.02 | 1.14 | 1.14 | 1.39 | 1.15 | 1.04 | -1.06 |
| fatty acid oxidation | 13 | -1.06 | 1.08 | -1.52 | 1.06 | 1.2 | -2.01 | -1.29 |
| alditol metabolic process | 16 | -1.18 | 1.1 | -1.05 | 1.11 | 1.01 | -1.33 | -1.11 |
| alditol catabolic process | 2 | 1.2 | 1.12 | -1.49 | 1.16 | 1.79 | -1.21 | -1.08 |
| allantoin biosynthetic process | 1 | 3.87 | -2.08 | -2.11 | -2.0 | -1.26 | -1.05 | -1.64 |
| removal of superoxide radicals | 2 | 1.69 | 1.17 | 1.01 | 1.09 | 1.11 | 1.41 | -1.08 |
| triglyceride biosynthetic process | 1 | -1.05 | 1.33 | 3.49 | 1.16 | 1.98 | 2.09 | 1.25 |
| triglyceride catabolic process | 2 | 1.6 | 1.03 | -1.14 | -1.01 | 1.9 | -1.45 | -1.11 |
| aromatic compound biosynthetic process | 24 | 1.66 | -1.07 | -1.13 | -1.07 | -1.14 | 1.23 | 1.24 |
| aromatic compound catabolic process | 8 | -1.06 | -1.01 | -1.07 | -1.04 | -1.14 | 1.17 | -1.07 |
| tryptophan catabolic process to kynurenine | 1 | 1.33 | -1.46 | -1.36 | -1.56 | -1.39 | 2.84 | -1.33 |
| 4-hydroxyproline metabolic process | 15 | -1.0 | 1.02 | -1.1 | -1.04 | -1.12 | 1.01 | -1.04 |
| D-amino acid catabolic process | 1 | 1.5 | 1.05 | 1.18 | 1.45 | -1.04 | 1.27 | -1.33 |
| beta-alanine metabolic process | 1 | 1.44 | -1.11 | -1.15 | 1.16 | -1.17 | -1.1 | -1.2 |
| beta-alanine biosynthetic process | 1 | 1.44 | -1.11 | -1.15 | 1.16 | -1.17 | -1.1 | -1.2 |
| L-methionine salvage from methylthioadenosine | 1 | 1.92 | 1.16 | 1.15 | 1.28 | 1.06 | 2.87 | -1.21 |
| peptidyl-proline hydroxylation | 15 | -1.0 | 1.02 | -1.1 | -1.04 | -1.12 | 1.01 | -1.04 |
| protein metabolic process | 1632 | -1.11 | -1.03 | 1.01 | 1.05 | 1.08 | -1.08 | -1.1 |
| arginine catabolic process to glutamate | 1 | -13.01 | 1.26 | -21.51 | 1.04 | 1.21 | -8.69 | 1.74 |
| arginine catabolic process to ornithine | 1 | 1.27 | -1.42 | -1.22 | -1.31 | -1.39 | 3.47 | -1.56 |
| glutamate catabolic process to 2-oxoglutarate | 2 | -1.13 | -1.34 | 1.15 | -1.41 | 1.89 | 2.28 | -2.14 |
| glycerol catabolic process | 2 | 1.2 | 1.12 | -1.49 | 1.16 | 1.79 | -1.21 | -1.08 |
| catechol-containing compound catabolic process | 1 | -8.98 | 1.77 | -2.05 | -1.02 | -2.08 | -14.22 | 1.22 |
| urea metabolic process | 1 | -31.89 | 1.77 | -20.72 | 1.41 | 1.0 | -12.66 | -3.5 |
| organophosphate metabolic process | 108 | 1.04 | -1.04 | 1.02 | 1.03 | 1.24 | 1.03 | -1.05 |
| GDP-mannose metabolic process | 2 | -2.16 | 1.21 | 1.23 | 1.27 | 1.35 | -2.29 | 1.03 |
| NAD metabolic process | 4 | 1.51 | 1.15 | -1.0 | -1.2 | 1.44 | 1.73 | 1.09 |
| choline metabolic process | 1 | 2.27 | 1.23 | -5.17 | -1.59 | -2.2 | -9.9 | 9.42 |
| protein-cysteine S-palmitoleyltransferase activity | 21 | -1.12 | 1.09 | 1.11 | 1.11 | 1.07 | -1.16 | -1.11 |
| protein-cysteine S-acyltransferase activity | 21 | -1.12 | 1.09 | 1.11 | 1.11 | 1.07 | -1.16 | -1.11 |
| Mo-molybdopterin cofactor metabolic process | 5 | 1.14 | 1.31 | 1.37 | 1.14 | 1.16 | -1.34 | 1.2 |
| B cell mediated immunity | 2 | -2.37 | 1.1 | 1.16 | -1.04 | 1.34 | -1.89 | 1.24 |
| cellular homeostasis | 118 | 1.06 | 1.01 | 1.08 | -1.01 | 1.03 | 1.15 | 1.0 |
| antimicrobial humoral response | 59 | 1.29 | -1.22 | -1.15 | -1.21 | 1.01 | 1.23 | 1.27 |
| antibacterial humoral response | 22 | 1.72 | -1.69 | -1.59 | -1.57 | -1.23 | 1.37 | 1.27 |
| antifungal humoral response | 8 | 4.25 | -1.39 | -1.37 | -1.5 | -1.09 | 3.13 | 1.41 |
| regulation of isoprenoid metabolic process | 2 | -8.29 | -1.83 | 1.21 | 1.4 | 1.08 | -7.35 | -3.84 |
| secondary metabolic process | 53 | 1.57 | -1.09 | -1.23 | -1.11 | -1.05 | 1.14 | 1.21 |
| cytoskeleton-dependent cytoplasmic transport, nurse cell to oocyte | 1 | -3.92 | 1.08 | 1.44 | 1.19 | 1.39 | -1.01 | -1.05 |
| polyol metabolic process | 18 | -1.04 | -1.02 | -1.13 | 1.01 | -1.03 | -1.13 | -1.13 |
| carboxylic acid metabolic process | 239 | -1.01 | -1.01 | -1.15 | -1.03 | 1.0 | 1.06 | -1.05 |
| proteasome core complex, alpha-subunit complex | 10 | 1.29 | -1.03 | 1.04 | 1.15 | 1.11 | -1.27 | -1.17 |
| NEDD8 activating enzyme activity | 3 | 1.18 | 1.04 | 1.17 | 1.22 | 1.25 | -1.07 | -1.14 |
| small conjugating protein-specific protease activity | 9 | 1.26 | 1.18 | 1.74 | 1.17 | 1.51 | 1.89 | 1.35 |
| NEDD8-specific protease activity | 1 | 2.27 | 1.0 | 1.29 | 1.04 | 1.1 | 1.44 | -1.06 |
| small conjugating protein ligase activity | 84 | 1.15 | 1.18 | 1.44 | 1.14 | 1.34 | 1.23 | 1.22 |
| NEDD8 ligase activity | 2 | -1.0 | -1.06 | 1.71 | -1.01 | 1.42 | -1.21 | -1.32 |
| SUMO ligase activity | 1 | 1.22 | 1.25 | 1.21 | 1.04 | 1.21 | 1.26 | 1.19 |
| procollagen-proline dioxygenase activity | 26 | -1.01 | -1.0 | -1.08 | -1.01 | -1.13 | -1.04 | -1.05 |
| tubulin N-acetyltransferase activity | 2 | 1.47 | -1.06 | 1.07 | -1.01 | 1.13 | 1.11 | 1.06 |
| oxygen sensor activity | 1 | -1.13 | -1.1 | -1.21 | -1.08 | -1.27 | -1.0 | -1.19 |
| stem cell maintenance | 50 | 1.1 | 1.12 | 1.31 | 1.12 | 1.35 | 1.12 | 1.22 |
| response to cadmium ion | 7 | -1.02 | 1.14 | 1.52 | 1.17 | 1.43 | -1.04 | 1.03 |
| isoprenoid binding | 10 | 1.91 | -4.39 | -4.6 | -2.0 | -1.48 | 1.68 | 1.12 |
| retinol binding | 1 | 1.33 | -27.02 | -29.08 | -25.07 | -1.99 | 8.32 | -2.83 |
| vitamin binding | 93 | -1.14 | -1.23 | -1.54 | -1.07 | -1.09 | -1.09 | -1.12 |
| rRNA binding | 11 | 1.14 | 1.1 | 1.07 | 1.22 | 1.14 | 1.03 | -1.12 |
| pyrimidine base biosynthetic process | 6 | 1.66 | -1.0 | 1.02 | -1.03 | -1.03 | 1.11 | 1.08 |
| uracil metabolic process | 1 | 1.44 | -1.11 | -1.15 | 1.16 | -1.17 | -1.1 | -1.2 |
| flagellum | 7 | 1.02 | 1.05 | 1.0 | 1.02 | -1.02 | 1.02 | 1.03 |
| organelle inner membrane | 160 | 1.06 | 1.05 | 1.17 | 1.05 | 1.11 | 1.38 | -1.22 |
| outer membrane | 27 | 1.24 | -1.02 | 1.15 | 1.09 | 1.2 | 1.39 | 1.01 |
| protein phosphatase regulator activity | 31 | 1.4 | -1.05 | 1.16 | 1.02 | 1.17 | 1.37 | 1.12 |
| pteridine metabolic process | 5 | 2.4 | -1.22 | -1.26 | -1.24 | -1.16 | -1.04 | 1.7 |
| kinesin binding | 7 | 1.13 | 1.16 | 1.23 | 1.15 | 1.17 | 1.39 | 1.05 |
| axon transport of mitochondrion | 3 | 1.06 | 1.09 | 1.22 | 1.23 | 1.2 | 1.22 | -1.08 |
| extrinsic to plasma membrane | 31 | 1.23 | 1.05 | 1.3 | 1.05 | 1.11 | 1.03 | 1.06 |
| extrinsic to membrane | 45 | 1.12 | 1.02 | 1.14 | 1.05 | 1.1 | 1.06 | 1.1 |
| kinase binding | 30 | 1.28 | 1.21 | 1.48 | 1.12 | 1.41 | 1.31 | 1.33 |
| protein kinase binding | 24 | 1.38 | 1.18 | 1.53 | 1.14 | 1.4 | 1.3 | 1.3 |
| phosphatase binding | 25 | 1.23 | -1.0 | 1.19 | 1.06 | 1.18 | 1.25 | 1.15 |
| protein phosphatase binding | 25 | 1.23 | -1.0 | 1.19 | 1.06 | 1.18 | 1.25 | 1.15 |
| protein domain specific binding | 37 | 1.41 | 1.06 | 1.28 | 1.05 | 1.2 | 1.48 | 1.26 |
| syntaxin binding | 3 | -1.34 | 1.02 | 1.2 | 1.13 | 1.16 | 1.09 | 1.32 |
| nuclear cyclin-dependent protein kinase holoenzyme complex | 7 | 1.06 | 1.09 | 1.22 | 1.21 | 1.41 | -1.23 | -1.12 |
| [pyruvate dehydrogenase (lipoamide)] phosphatase regulator activity | 1 | 1.97 | 1.3 | 1.52 | -1.19 | 1.01 | 1.99 | 1.65 |
| mitochondrial pyruvate dehydrogenase (lipoamide) phosphatase complex | 1 | 1.97 | 1.3 | 1.52 | -1.19 | 1.01 | 1.99 | 1.65 |
| lipid storage | 14 | 1.31 | 1.0 | 1.02 | 1.14 | 1.38 | 1.02 | 1.12 |
| peptidyl-arginine methylation, to asymmetrical-dimethyl arginine | 3 | 1.19 | 1.12 | 1.45 | 1.13 | 1.36 | 1.2 | 1.09 |
| second-messenger-mediated signaling | 22 | 1.04 | 1.12 | 1.05 | -1.04 | -1.06 | 1.14 | 1.04 |
| cAMP-mediated signaling | 12 | 1.11 | 1.19 | 1.15 | -1.07 | 1.01 | 1.19 | 1.11 |
| cGMP-mediated signaling | 1 | -1.03 | -1.03 | -1.03 | -1.11 | -1.15 | 1.16 | -1.29 |
| cyclic-nucleotide-mediated signaling | 13 | 1.1 | 1.17 | 1.14 | -1.08 | -1.0 | 1.18 | 1.08 |
| phosphatidylinositol-mediated signaling | 4 | -1.13 | 1.15 | 1.6 | 1.12 | 1.84 | 1.36 | 1.05 |
| inositol phosphate-mediated signaling | 4 | 1.12 | 1.13 | 1.06 | 1.07 | 1.06 | 1.07 | 1.09 |
| inositol lipid-mediated signaling | 4 | -1.13 | 1.15 | 1.6 | 1.12 | 1.84 | 1.36 | 1.05 |
| protein neddylation | 3 | 1.49 | 1.06 | 1.55 | 1.26 | 1.54 | 1.44 | -1.06 |
| SUMO activating enzyme activity | 2 | 1.33 | 1.09 | 1.29 | 1.43 | 1.67 | 1.2 | -1.13 |
| SMT3-dependent protein catabolic process | 2 | 1.33 | 1.09 | 1.29 | 1.43 | 1.67 | 1.2 | -1.13 |
| sexual reproduction | 596 | 1.13 | 1.1 | 1.22 | 1.05 | 1.25 | 1.21 | 1.19 |
| translesion synthesis | 4 | 1.17 | 1.02 | 1.32 | 1.08 | 1.28 | 1.43 | 1.07 |
| negative regulation of anti-apoptosis | 1 | -1.96 | -1.03 | 2.81 | 1.35 | 1.53 | -2.84 | -1.09 |
| charged-tRNA amino acid modification | 1 | 1.38 | 1.09 | 1.78 | 1.2 | 1.53 | 2.7 | -1.25 |
| septate junction assembly | 24 | 2.8 | -1.27 | 1.3 | -1.27 | 1.2 | 2.89 | 1.48 |
| diacylglycerol binding | 24 | 1.82 | -1.05 | 1.33 | -1.08 | 1.26 | 1.8 | 1.26 |
| heme binding | 133 | -1.06 | -1.16 | -1.25 | -1.06 | -1.02 | -1.05 | -1.12 |
| spinal cord development | 1 | 6.63 | -1.06 | 1.58 | -1.04 | 1.02 | 3.09 | 1.27 |
| spinal cord patterning | 1 | 6.63 | -1.06 | 1.58 | -1.04 | 1.02 | 3.09 | 1.27 |
| spinal cord dorsal/ventral patterning | 1 | 6.63 | -1.06 | 1.58 | -1.04 | 1.02 | 3.09 | 1.27 |
| telencephalon development | 2 | -1.33 | -1.42 | -1.06 | 1.28 | 1.22 | -1.65 | -2.16 |
| medulla oblongata development | 2 | -1.31 | 1.27 | 1.45 | 1.03 | 1.47 | -1.24 | 1.48 |
| central nervous system morphogenesis | 3 | -1.29 | 1.06 | 1.02 | -1.08 | -1.1 | -1.62 | 1.35 |
| central nervous system formation | 1 | 2.75 | -1.14 | -1.16 | -1.07 | -1.18 | 1.11 | 1.67 |
| nerve development | 4 | 1.97 | -1.1 | 1.31 | -1.24 | 1.47 | 2.5 | 1.71 |
| nerve maturation | 3 | 3.11 | -1.4 | 1.27 | -1.39 | 1.24 | 4.46 | 1.69 |
| developmental maturation | 131 | 1.17 | 1.08 | 1.28 | 1.04 | 1.26 | 1.43 | 1.24 |
| glial cell fate commitment | 3 | 1.44 | 1.3 | 1.68 | 1.12 | 1.63 | 1.5 | 1.59 |
| glial cell development | 20 | 2.6 | -1.18 | 1.22 | -1.13 | 1.22 | 1.95 | 1.32 |
| negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning | 1 | 6.63 | -1.06 | 1.58 | -1.04 | 1.02 | 3.09 | 1.27 |
| BMP signaling pathway involved in spinal cord dorsal/ventral patterning | 1 | 6.63 | -1.06 | 1.58 | -1.04 | 1.02 | 3.09 | 1.27 |
| central nervous system projection neuron axonogenesis | 2 | -1.33 | -1.42 | -1.06 | 1.28 | 1.22 | -1.65 | -2.16 |
| central nervous system neuron differentiation | 4 | -1.09 | -1.06 | 1.3 | 1.23 | 1.45 | 1.03 | -1.16 |
| central nervous system neuron development | 4 | -1.09 | -1.06 | 1.3 | 1.23 | 1.45 | 1.03 | -1.16 |
| central nervous system neuron axonogenesis | 3 | -1.36 | -1.18 | 1.1 | 1.26 | 1.32 | -1.27 | -1.53 |
| anterior commissure morphogenesis | 2 | -1.33 | -1.42 | -1.06 | 1.28 | 1.22 | -1.65 | -2.16 |
| neurogenesis | 1058 | 1.13 | 1.07 | 1.21 | 1.08 | 1.23 | 1.18 | 1.13 |
| regulation of rhodopsin mediated signaling pathway | 13 | 1.3 | 1.02 | 1.13 | 1.05 | 1.11 | 1.08 | 1.05 |
| cell cycle process | 518 | 1.19 | 1.1 | 1.22 | 1.11 | 1.27 | 1.19 | 1.09 |
| cell cycle phase | 443 | 1.2 | 1.1 | 1.22 | 1.11 | 1.26 | 1.19 | 1.09 |
| molting cycle process | 32 | 1.34 | -1.12 | -1.05 | -1.15 | -1.08 | 1.23 | 1.26 |
| regulation of cell-cell adhesion | 11 | 1.5 | 1.16 | 1.29 | 1.11 | 1.45 | 1.61 | 1.51 |
| negative regulation of cell-cell adhesion | 8 | 1.85 | 1.18 | 1.27 | 1.06 | 1.22 | 1.87 | 1.48 |
| positive regulation of cell-cell adhesion | 2 | 1.24 | 1.07 | 1.55 | 1.24 | 2.26 | 1.04 | 1.52 |
| circadian sleep/wake cycle process | 17 | -1.14 | 1.18 | 1.14 | -1.02 | 1.04 | 1.12 | 1.16 |
| cellular component disassembly | 27 | -1.1 | 1.18 | 1.3 | 1.14 | 1.29 | 1.09 | 1.04 |
| cellular process involved in reproduction in multicellular organism | 70 | 1.23 | 1.09 | 1.22 | 1.06 | 1.2 | 1.11 | 1.16 |
| reproductive process | 803 | 1.12 | 1.07 | 1.17 | 1.04 | 1.2 | 1.16 | 1.15 |
| viral reproductive process | 3 | -1.12 | 1.07 | 1.27 | -1.16 | -1.11 | 1.69 | 1.21 |
| chaeta development | 83 | 1.13 | 1.09 | 1.31 | 1.05 | 1.21 | 1.1 | 1.15 |
| digestive system process | 1 | 1.59 | -1.18 | -1.24 | -1.07 | -1.14 | 4.1 | -1.27 |
| ovulation cycle process | 1 | -1.49 | 2.58 | -1.26 | -1.23 | -1.23 | -1.38 | 1.24 |
| regulation of anatomical structure morphogenesis | 194 | 1.14 | 1.09 | 1.33 | 1.05 | 1.32 | 1.28 | 1.21 |
| regulation of cell morphogenesis | 139 | 1.13 | 1.12 | 1.38 | 1.05 | 1.36 | 1.36 | 1.25 |
| establishment of proximal/distal cell polarity | 3 | 1.72 | -1.01 | 1.03 | -1.08 | 1.97 | 1.34 | 1.24 |
| multicellular organism adhesion | 11 | -1.06 | 1.01 | -1.06 | -1.01 | -1.12 | -1.06 | -1.12 |
| multicellular organism adhesion to substrate | 11 | -1.06 | 1.01 | -1.06 | -1.01 | -1.12 | -1.06 | -1.12 |
| biological adhesion | 172 | 1.2 | -1.06 | 1.06 | -1.06 | 1.19 | 1.34 | 1.23 |
| dormancy process | 1 | -1.14 | 1.2 | 1.7 | 1.09 | 2.04 | 1.39 | 1.74 |
| gland morphogenesis | 122 | 1.06 | 1.0 | 1.13 | 1.04 | 1.25 | 1.23 | 1.18 |
| ribonucleoprotein complex biogenesis | 81 | 1.26 | 1.15 | 1.26 | 1.26 | 1.3 | 1.14 | 1.07 |
| DNA strand elongation | 3 | -1.21 | -1.08 | 1.4 | 1.14 | 1.61 | -1.08 | -1.25 |
| extracellular matrix disassembly | 3 | -2.47 | 1.3 | 1.22 | 1.11 | 1.41 | -1.09 | 1.18 |
| ribonucleoprotein complex assembly | 33 | 1.24 | 1.13 | 1.29 | 1.19 | 1.29 | 1.24 | 1.1 |
| proteasome accessory complex | 25 | 1.35 | 1.08 | 1.25 | 1.18 | 1.19 | 1.07 | 1.06 |
| cytosolic ribosome | 83 | 1.38 | 1.02 | 1.1 | 1.01 | 1.14 | 1.19 | 1.07 |
| passive transmembrane transporter activity | 217 | 1.02 | 1.0 | -1.01 | -1.05 | -1.07 | 1.03 | -1.01 |
| active transmembrane transporter activity | 330 | -1.26 | -1.06 | -1.28 | -1.12 | -1.06 | 1.14 | -1.11 |
| substrate-specific transmembrane transporter activity | 609 | -1.14 | -1.03 | -1.14 | -1.09 | -1.07 | 1.09 | -1.07 |
| potassium ion symporter activity | 5 | -1.79 | -1.85 | -2.57 | -1.19 | 1.65 | 1.54 | 1.09 |
| potassium ion antiporter activity | 5 | -1.42 | 1.17 | -1.18 | 1.1 | -1.14 | -1.59 | -1.02 |
| copper-transporting ATPase activity | 1 | -2.22 | 1.33 | 3.02 | 1.07 | -1.16 | -1.12 | 1.08 |
| wide pore channel activity | 7 | -1.65 | -1.07 | -1.04 | -1.07 | -1.24 | -1.19 | 1.29 |
| voltage-gated channel activity | 44 | 1.07 | -1.01 | 1.02 | -1.09 | -1.09 | 1.15 | -1.09 |
| mechanically gated channel activity | 1 | -1.05 | 1.0 | -1.06 | -1.03 | -1.14 | 1.01 | 1.95 |
| ligand-gated channel activity | 108 | 1.02 | 1.0 | -1.0 | -1.08 | -1.14 | 1.0 | -1.02 |
| gated channel activity | 149 | 1.05 | 1.0 | 1.02 | -1.05 | -1.09 | 1.03 | -1.02 |
| substrate-specific channel activity | 210 | 1.03 | 1.0 | -1.0 | -1.05 | -1.07 | 1.04 | -1.01 |
| ion gated channel activity | 3 | -1.25 | -1.55 | 1.25 | 1.49 | -1.39 | -1.46 | -1.49 |
| voltage-gated cation channel activity | 35 | 1.13 | -1.03 | 1.01 | -1.13 | -1.12 | 1.09 | -1.13 |
| macromolecule transmembrane transporter activity | 33 | 1.03 | 1.14 | 1.21 | 1.1 | 1.12 | 1.16 | -1.01 |
| inorganic cation transmembrane transporter activity | 219 | -1.22 | -1.02 | -1.2 | -1.15 | -1.13 | 1.1 | -1.15 |
| substrate-specific transporter activity | 669 | -1.12 | -1.02 | -1.12 | -1.08 | -1.06 | 1.1 | -1.06 |
| regulation of transmembrane transporter activity | 7 | 1.44 | -1.04 | 1.16 | -1.02 | 1.15 | 1.52 | 1.13 |
| neurotransmitter receptor activity | 51 | 1.09 | -1.06 | 1.03 | -1.05 | -1.03 | 1.12 | -1.01 |
| cellular response to amino acid stimulus | 4 | 1.59 | -1.01 | 1.38 | 1.29 | 1.34 | 1.2 | -1.18 |
| termination of signal transduction | 9 | 1.61 | 1.14 | 1.6 | 1.12 | 1.38 | 1.47 | 1.34 |
| paracrine signaling | 3 | -1.1 | 1.05 | 1.09 | 1.11 | 1.21 | -1.07 | 1.08 |
| cellular response to stress | 329 | 1.18 | 1.1 | 1.23 | 1.1 | 1.28 | 1.2 | 1.12 |
| signaling | 1219 | 1.14 | 1.03 | 1.15 | 1.03 | 1.17 | 1.18 | 1.11 |
| cellular response to chemical stimulus | 143 | 1.16 | 1.06 | 1.21 | 1.05 | 1.2 | 1.2 | 1.12 |
| regulation of signaling | 409 | 1.15 | 1.12 | 1.28 | 1.06 | 1.24 | 1.27 | 1.22 |
| cellular response to mechanical stimulus | 1 | -1.7 | -15.86 | -19.15 | -19.05 | -4.94 | 3.57 | 1.25 |
| positive regulation of signaling | 85 | 1.13 | 1.11 | 1.24 | 1.05 | 1.27 | 1.19 | 1.24 |
| negative regulation of signaling | 149 | 1.2 | 1.1 | 1.31 | 1.06 | 1.27 | 1.32 | 1.25 |
| adaptation of signaling pathway | 4 | -1.08 | 1.14 | 1.07 | 1.08 | 1.11 | -1.0 | -1.04 |
| signal release | 101 | -1.07 | 1.04 | 1.24 | 1.13 | 1.26 | 1.1 | 1.03 |
| cellular cation homeostasis | 29 | 1.01 | 1.02 | 1.39 | -1.05 | -1.07 | 1.29 | 1.04 |
| cellular monovalent inorganic cation homeostasis | 6 | -1.56 | 1.28 | 1.81 | 1.0 | 1.29 | 1.81 | 1.32 |
| cellular divalent inorganic cation homeostasis | 11 | 1.74 | 1.02 | 1.47 | -1.09 | -1.04 | 1.57 | 1.16 |
| TRAPP complex | 3 | 1.9 | 1.04 | 1.48 | 1.37 | 1.5 | 1.53 | 1.03 |
| maintenance of cell polarity | 12 | -1.07 | 1.19 | 1.56 | 1.06 | 1.35 | 1.28 | 1.22 |
| CCR4-NOT complex | 7 | 1.25 | 1.15 | 1.58 | 1.15 | 1.35 | 1.67 | 1.21 |
| myofibril | 29 | 2.16 | -1.42 | -1.09 | -1.18 | 1.24 | 1.83 | -1.17 |
| sarcomere | 28 | 2.2 | -1.44 | -1.09 | -1.19 | 1.26 | 1.88 | -1.18 |
| Z disc | 19 | 1.85 | -1.41 | -1.18 | -1.11 | 1.31 | 1.7 | -1.14 |
| extracellular matrix binding | 1 | -1.18 | -1.04 | -1.07 | -1.26 | 1.04 | 1.06 | 5.07 |
| lamellipodium | 1 | -1.44 | 1.23 | 1.5 | 1.23 | 1.55 | 1.34 | 1.29 |
| actin filament-based process | 163 | 1.17 | 1.03 | 1.35 | -1.03 | 1.3 | 1.31 | 1.17 |
| cell projection organization | 408 | 1.12 | 1.08 | 1.23 | 1.04 | 1.24 | 1.23 | 1.23 |
| cell projection assembly | 64 | -1.14 | 1.06 | 1.16 | 1.07 | 1.16 | 1.01 | 1.05 |
| lamellipodium assembly | 7 | 1.05 | 1.14 | 1.49 | 1.08 | 1.51 | 1.25 | 1.41 |
| microvillar actin bundle assembly | 1 | -2.22 | -1.56 | -1.05 | -1.91 | -2.01 | -2.21 | 7.0 |
| actin filament reorganization involved in cell cycle | 3 | -1.25 | 1.17 | 1.3 | 1.13 | 1.1 | -1.42 | -1.1 |
| contractile actin filament bundle assembly | 1 | 2.13 | 1.0 | -1.14 | -1.14 | 1.0 | 1.74 | 1.58 |
| cell junction | 105 | 1.39 | -1.0 | 1.25 | -1.05 | 1.25 | 1.62 | 1.33 |
| cell-substrate junction | 22 | 1.32 | 1.08 | 1.37 | 1.01 | 1.39 | 1.3 | 1.26 |
| desmosome | 1 | 1.71 | 1.23 | 1.34 | 1.26 | 1.67 | -1.28 | -1.04 |
| L-malate dehydrogenase activity | 2 | -1.06 | -1.01 | 1.02 | -1.02 | -1.07 | 1.05 | -1.04 |
| mitochondrial crista | 1 | 1.79 | 1.07 | 1.12 | 1.18 | 1.43 | 1.54 | -1.19 |
| mitochondrial tricarboxylic acid cycle enzyme complex | 5 | 1.01 | -1.02 | 1.12 | -1.1 | 1.02 | 1.43 | -1.32 |
| regulation of mitotic metaphase/anaphase transition | 22 | 1.12 | 1.08 | 1.16 | 1.15 | 1.3 | 1.05 | -1.0 |
| protein repair | 1 | 2.93 | 1.03 | 1.2 | 1.19 | 1.05 | 1.97 | 1.02 |
| hemopoiesis | 41 | 1.12 | 1.09 | 1.4 | 1.05 | 1.34 | 1.25 | 1.41 |
| regulation of endocytosis | 16 | 1.17 | 1.02 | 1.49 | 1.1 | 1.26 | 1.72 | 1.22 |
| water homeostasis | 2 | -1.06 | -1.23 | 1.57 | -1.02 | -2.03 | 1.99 | 1.2 |
| regulation of Wnt receptor signaling pathway | 46 | 1.23 | 1.14 | 1.25 | 1.09 | 1.25 | 1.37 | 1.32 |
| membrane coat | 25 | -1.02 | 1.18 | 1.24 | 1.13 | 1.15 | 1.19 | 1.33 |
| clathrin coat | 12 | -1.14 | 1.2 | 1.34 | 1.14 | 1.23 | 1.23 | 1.26 |
| vesicle coat | 14 | -1.23 | 1.19 | 1.13 | 1.12 | 1.05 | 1.13 | 1.37 |
| AP-3 adaptor complex | 1 | 1.85 | 1.2 | 1.21 | 1.06 | 1.07 | 1.4 | 1.37 |
| clathrin vesicle coat | 4 | -1.28 | 1.18 | 1.41 | 1.09 | 1.26 | 1.54 | 1.36 |
| COPII vesicle coat | 1 | 1.02 | 1.56 | 2.15 | 1.06 | 2.07 | 2.36 | 2.5 |
| clathrin coat of synaptic vesicle | 1 | -1.18 | 1.22 | 1.63 | -1.04 | 1.48 | 2.58 | 1.85 |
| clathrin coat of trans-Golgi network vesicle | 3 | -1.23 | 1.16 | 1.44 | 1.05 | 1.28 | 1.6 | 1.31 |
| clathrin adaptor complex | 9 | -1.11 | 1.2 | 1.31 | 1.16 | 1.2 | 1.14 | 1.2 |
| clathrin coat of coated pit | 2 | -1.13 | 1.16 | 1.51 | 1.02 | 1.41 | 1.68 | 1.45 |
| coated vesicle | 76 | 1.02 | 1.11 | 1.19 | 1.09 | 1.21 | 1.14 | 1.2 |
| clathrin-coated vesicle | 62 | 1.02 | 1.11 | 1.23 | 1.08 | 1.25 | 1.15 | 1.16 |
| trans-Golgi network transport vesicle | 6 | -1.41 | 1.21 | 1.34 | 1.14 | 1.2 | 1.25 | 1.25 |
| manganese ion binding | 18 | -1.13 | 1.08 | -1.01 | 1.1 | 1.02 | -1.14 | -1.09 |
| sphingolipid biosynthetic process | 7 | -1.12 | 1.12 | 1.25 | -1.02 | 1.07 | -1.09 | 1.53 |
| sphingolipid catabolic process | 7 | -6.83 | -3.3 | -2.99 | 1.5 | -1.08 | -6.52 | -5.81 |
| molybdenum ion binding | 3 | -1.62 | 1.26 | -1.41 | 1.36 | -1.14 | -1.63 | -1.18 |
| cell differentiation | 1609 | 1.15 | 1.07 | 1.19 | 1.06 | 1.21 | 1.19 | 1.12 |
| regulation of cell adhesion | 16 | 1.49 | 1.13 | 1.37 | 1.1 | 1.52 | 1.6 | 1.42 |
| protein xylosyltransferase activity | 1 | 1.17 | 1.3 | 1.39 | 1.35 | 1.31 | 1.22 | 1.24 |
| receptor signaling complex scaffold activity | 2 | 1.19 | 1.14 | 1.28 | -1.02 | 1.24 | 1.13 | 1.22 |
| regulation of proteolysis | 37 | 1.34 | 1.07 | 1.14 | 1.08 | 1.15 | 1.29 | 1.22 |
| protein catabolic process | 123 | 1.15 | 1.11 | 1.28 | 1.14 | 1.26 | 1.07 | 1.08 |
| PDZ domain binding | 5 | 1.3 | 1.06 | 1.19 | -1.03 | 1.16 | 1.22 | 1.15 |
| proteoglycan biosynthetic process | 17 | 1.25 | 1.14 | 1.21 | 1.06 | 1.15 | 1.25 | 1.53 |
| pyridoxal phosphate binding | 40 | 1.01 | 1.02 | -1.27 | -1.04 | -1.03 | -1.02 | 1.04 |
| integral to Golgi membrane | 5 | 1.35 | 1.04 | -1.19 | 1.04 | 1.29 | 1.43 | 1.52 |
| filopodium | 4 | 1.02 | 1.09 | 1.65 | -1.04 | 1.56 | 1.24 | 1.53 |
| integral to endoplasmic reticulum membrane | 10 | -1.46 | 1.11 | 1.14 | 1.2 | 1.05 | -1.72 | -1.17 |
| positive regulation of Wnt receptor signaling pathway | 14 | 1.4 | 1.13 | 1.25 | 1.11 | 1.22 | 1.24 | 1.36 |
| negative regulation of Wnt receptor signaling pathway | 21 | 1.2 | 1.09 | 1.17 | 1.1 | 1.24 | 1.55 | 1.3 |
| neuron differentiation | 458 | 1.15 | 1.09 | 1.26 | 1.04 | 1.27 | 1.27 | 1.24 |
| extracellular matrix organization | 25 | -1.06 | 1.03 | 1.16 | -1.04 | 1.09 | -1.05 | 1.22 |
| heparan sulfate proteoglycan metabolic process | 12 | 1.32 | 1.18 | 1.4 | 1.15 | 1.27 | 1.3 | 1.26 |
| heparin metabolic process | 2 | 1.46 | 1.48 | 1.74 | 1.25 | 2.09 | 1.65 | 1.44 |
| glycosaminoglycan metabolic process | 24 | 1.12 | -1.18 | -1.02 | 1.02 | -1.21 | -1.06 | 1.02 |
| chondroitin sulfate metabolic process | 7 | 1.31 | 1.13 | 1.16 | 1.13 | 1.24 | 1.3 | 1.08 |
| chondroitin sulfate biosynthetic process | 7 | 1.31 | 1.13 | 1.16 | 1.13 | 1.24 | 1.3 | 1.08 |
| heparin biosynthetic process | 2 | 1.46 | 1.48 | 1.74 | 1.25 | 2.09 | 1.65 | 1.44 |
| semaphorin receptor binding | 2 | -1.18 | -1.21 | 1.16 | -1.17 | 1.87 | 1.3 | 1.34 |
| lipoprotein particle receptor activity | 11 | 1.16 | 1.12 | 1.24 | -1.03 | -1.04 | 1.29 | 1.1 |
| enzyme regulator activity | 311 | 1.18 | -1.04 | 1.06 | -1.06 | 1.1 | 1.22 | 1.12 |
| female sex determination | 10 | 1.73 | 1.13 | 1.2 | 1.13 | 1.18 | 1.32 | 1.12 |
| male sex determination | 2 | 2.22 | 1.29 | 1.1 | 1.16 | 1.11 | -1.15 | 1.06 |
| myofibril assembly | 27 | 2.23 | -1.17 | 1.24 | -1.18 | 1.21 | 1.85 | 1.04 |
| skeletal muscle thin filament assembly | 2 | 3.07 | -1.51 | -1.03 | -1.18 | 1.0 | 1.63 | -1.62 |
| skeletal muscle myosin thick filament assembly | 4 | 2.6 | -1.28 | 1.1 | -1.18 | 1.07 | 1.9 | -1.04 |
| polysaccharide binding | 116 | -1.88 | -1.54 | -1.66 | -1.39 | -1.53 | -1.88 | 1.19 |
| guanylate cyclase regulator activity | 1 | 4.81 | -1.03 | -1.02 | 1.21 | 1.81 | 1.05 | 1.23 |
| guanylate cyclase activator activity | 1 | 4.81 | -1.03 | -1.02 | 1.21 | 1.81 | 1.05 | 1.23 |
| lipid modification | 43 | 1.21 | 1.09 | -1.03 | 1.03 | 1.31 | 1.05 | 1.06 |
| lipid glycosylation | 1 | 1.27 | 1.12 | -1.0 | 1.34 | 1.08 | 1.05 | -1.38 |
| entry into host cell | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| chromosome condensation | 38 | 1.16 | 1.12 | 1.26 | 1.12 | 1.35 | 1.24 | 1.15 |
| apoptotic nuclear change | 3 | 1.86 | 1.09 | 1.48 | 1.14 | 1.43 | 1.6 | 1.67 |
| glyoxylate reductase (NADP) activity | 2 | -4.26 | 1.85 | -1.64 | 1.04 | -1.41 | -1.97 | -1.08 |
| 5-formyltetrahydrofolate cyclo-ligase activity | 2 | -1.02 | 1.01 | 1.07 | 1.18 | 1.07 | 1.18 | -1.17 |
| clathrin binding | 3 | -1.33 | 1.27 | 1.39 | 1.12 | 1.36 | 1.07 | 1.24 |
| integral to synaptic vesicle membrane | 3 | 1.11 | 1.05 | 1.2 | 1.05 | 1.1 | 1.09 | 1.09 |
| dynein complex | 40 | -1.02 | 1.05 | 1.06 | 1.02 | -1.01 | 1.01 | -1.05 |
| outer membrane-bounded periplasmic space | 9 | 1.87 | -1.07 | 1.01 | -1.1 | -1.04 | 1.24 | -1.01 |
| protein phosphatase 4 complex | 1 | -1.33 | 1.33 | 1.64 | 1.24 | 1.24 | 1.44 | 1.37 |
| protein serine/threonine kinase inhibitor activity | 4 | 1.69 | 1.03 | 1.25 | 1.04 | 1.29 | 1.67 | 1.38 |
| protein tyrosine kinase inhibitor activity | 1 | 1.36 | 1.39 | 1.33 | -1.1 | 1.25 | 1.42 | 1.59 |
| protein kinase activator activity | 5 | 1.59 | 1.12 | 1.23 | 1.13 | 1.26 | 1.27 | 1.08 |
| protein tyrosine kinase activator activity | 3 | 1.95 | 1.11 | 1.44 | 1.07 | 1.38 | 1.82 | 1.28 |
| cholesterol transport | 1 | 2.58 | -1.39 | 1.67 | -1.07 | 1.62 | 11.34 | 1.83 |
| positive regulation of cell growth | 16 | 1.03 | 1.18 | 1.5 | 1.1 | 1.49 | 1.39 | 1.32 |
| negative regulation of cell growth | 9 | 1.27 | 1.11 | 1.45 | 1.06 | 1.45 | 1.67 | 1.24 |
| external encapsulating structure | 31 | 1.33 | -1.01 | -1.0 | 1.0 | -1.04 | -1.02 | -1.02 |
| cell envelope | 11 | 1.96 | -1.06 | 1.02 | -1.08 | -1.01 | 1.22 | 1.03 |
| junctional membrane complex | 1 | 2.03 | -1.16 | -1.04 | -1.08 | -1.05 | 1.25 | 1.07 |
| sperm motility | 9 | -1.26 | 1.06 | 1.05 | 1.04 | 1.04 | -1.0 | -1.05 |
| stabilization of membrane potential | 2 | 1.77 | 1.24 | 2.44 | -1.13 | 1.42 | 2.96 | 1.8 |
| DNA damage response, signal transduction by p53 class mediator | 2 | 1.7 | 1.37 | 1.03 | 1.24 | 1.62 | 1.04 | 1.2 |
| cyclin binding | 6 | 1.08 | 1.2 | 1.43 | 1.19 | 1.34 | -1.04 | 1.0 |
| regulation of cell migration | 10 | 2.17 | -1.1 | 1.06 | -1.11 | 1.21 | 1.34 | 1.71 |
| negative regulation of cell migration | 2 | 2.1 | 1.1 | 1.42 | -1.08 | 1.39 | 1.72 | 1.26 |
| DNA polymerase processivity factor activity | 2 | -1.29 | 1.09 | 1.33 | 1.22 | 1.3 | -1.6 | -1.28 |
| iron-responsive element binding | 2 | 1.36 | 1.24 | 1.32 | -1.06 | 1.1 | 1.69 | 1.16 |
| translation repressor activity | 10 | 1.56 | 1.06 | 1.57 | 1.02 | 1.3 | 1.21 | 1.77 |
| ligand-dependent nuclear receptor transcription coactivator activity | 3 | 1.32 | 1.16 | 1.48 | 1.23 | 1.62 | 1.33 | 1.22 |
| chorion-containing eggshell pattern formation | 8 | 1.93 | 1.01 | 1.22 | 1.01 | 1.3 | 1.72 | 1.37 |
| sperm mitochondrion organization | 5 | 1.07 | 1.08 | 1.53 | 1.1 | 1.13 | 1.48 | -1.14 |
| symbiosis, encompassing mutualism through parasitism | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| membrane disassembly | 3 | 1.03 | 1.27 | 1.04 | 1.1 | -1.14 | -1.28 | 1.1 |
| defecation | 1 | 1.59 | -1.18 | -1.24 | -1.07 | -1.14 | 4.1 | -1.27 |
| production of siRNA involved in RNA interference | 8 | 1.41 | 1.15 | 1.49 | 1.04 | 1.38 | 1.34 | 1.32 |
| targeting of mRNA for destruction involved in RNA interference | 4 | 1.09 | 1.13 | 1.5 | 1.03 | 1.41 | 1.21 | 1.37 |
| axon | 64 | 1.18 | 1.07 | 1.23 | -1.05 | 1.23 | 1.51 | 1.31 |
| dendrite | 39 | 1.09 | 1.05 | 1.22 | 1.05 | 1.15 | 1.21 | 1.12 |
| growth cone | 8 | 1.23 | 1.14 | 1.47 | 1.06 | 1.36 | 1.38 | 1.21 |
| site of polarized growth | 8 | 1.23 | 1.14 | 1.47 | 1.06 | 1.36 | 1.38 | 1.21 |
| sleep | 24 | 1.02 | 1.2 | 1.17 | 1.06 | 1.06 | 1.1 | 1.2 |
| peristalsis | 1 | 1.69 | 1.43 | 1.3 | 1.01 | 1.12 | 1.63 | -1.01 |
| ER-associated protein catabolic process | 5 | 1.01 | 1.28 | -1.09 | 1.11 | -1.01 | -1.06 | 1.46 |
| nuclear migration along microtubule | 1 | 2.24 | -1.02 | -1.01 | 1.05 | 1.3 | 1.83 | 1.39 |
| actin cap | 1 | 1.67 | 1.22 | 2.78 | 1.16 | 2.89 | 2.99 | 1.93 |
| tRNA methylation | 2 | 1.26 | 1.07 | -1.2 | 1.26 | 1.13 | -1.1 | -1.17 |
| midbody | 13 | 1.0 | 1.04 | 1.38 | 1.14 | 1.39 | -1.01 | 1.0 |
| fatty acid elongation | 3 | 1.23 | -2.04 | -1.81 | -2.67 | -2.07 | -1.23 | 3.6 |
| ankyrin binding | 2 | -1.35 | 1.17 | 1.37 | -1.02 | 1.05 | 1.36 | 1.56 |
| spectrin binding | 2 | 2.03 | 1.57 | 2.46 | 1.05 | 2.04 | 3.15 | 3.44 |
| BMP signaling pathway | 11 | 1.35 | 1.07 | 1.35 | 1.04 | 1.18 | 1.02 | 1.19 |
| regulation of BMP signaling pathway | 13 | -1.0 | 1.15 | 1.18 | 1.21 | 1.31 | -1.14 | 1.09 |
| positive regulation of transforming growth factor beta receptor signaling pathway | 1 | 2.13 | -1.08 | 1.11 | -1.24 | -1.05 | 1.06 | 1.61 |
| negative regulation of transforming growth factor beta receptor signaling pathway | 6 | 1.52 | 1.09 | 1.44 | -1.04 | 1.19 | 1.36 | 1.28 |
| positive regulation of BMP signaling pathway | 2 | -1.03 | 1.19 | -1.01 | 1.09 | 1.24 | -1.15 | 1.05 |
| negative regulation of BMP signaling pathway | 6 | -1.26 | 1.23 | 1.3 | 1.36 | 1.55 | -1.44 | 1.09 |
| snoRNA binding | 5 | 1.02 | 1.09 | 1.38 | 1.3 | 1.36 | -1.14 | -1.0 |
| regulation of axon extension | 7 | -1.17 | 1.19 | 1.37 | 1.14 | 1.29 | 1.18 | 1.05 |
| negative regulation of axon extension | 3 | -1.29 | 1.15 | 1.64 | 1.15 | 1.3 | -1.22 | -1.1 |
| intracellular steroid hormone receptor signaling pathway | 8 | 1.35 | 1.22 | 1.56 | 1.09 | 1.48 | 1.56 | 1.37 |
| intracellular receptor mediated signaling pathway | 12 | 1.17 | 1.24 | 1.48 | 1.1 | 1.54 | 1.37 | 1.29 |
| dihydrolipoamide S-acyltransferase activity | 2 | 1.46 | 1.31 | 1.81 | -1.03 | 1.35 | 2.15 | 1.15 |
| male genitalia development | 16 | 1.31 | 1.09 | 1.3 | 1.05 | 1.18 | 1.05 | 1.22 |
| female genitalia development | 6 | 1.5 | 1.12 | 1.14 | 1.04 | 1.07 | 1.14 | 1.13 |
| receptor regulator activity | 3 | 1.55 | 1.1 | 1.06 | 1.1 | 1.12 | 1.11 | 1.09 |
| receptor activator activity | 1 | 1.23 | 1.27 | 1.15 | 1.19 | 1.2 | 1.15 | 1.07 |
| receptor inhibitor activity | 2 | 1.75 | 1.03 | 1.01 | 1.06 | 1.08 | 1.1 | 1.1 |
| cyclic nucleotide binding | 2 | -1.42 | -1.32 | -1.55 | -1.36 | -1.51 | 4.6 | 1.1 |
| adenyl nucleotide binding | 654 | 1.06 | 1.1 | 1.17 | 1.09 | 1.22 | 1.18 | 1.09 |
| rRNA modification guide activity | 2 | 1.38 | 1.09 | 1.14 | 1.35 | 1.29 | -1.02 | -1.04 |
| RNA pseudouridylation guide activity | 2 | 1.38 | 1.09 | 1.14 | 1.35 | 1.29 | -1.02 | -1.04 |
| rRNA pseudouridylation guide activity | 2 | 1.38 | 1.09 | 1.14 | 1.35 | 1.29 | -1.02 | -1.04 |
| phosphatidyltransferase activity | 1 | 1.31 | 1.29 | 1.34 | 1.27 | 1.36 | 1.03 | -1.44 |
| nuclear body organization | 2 | -1.19 | 1.31 | 1.94 | 1.17 | 1.95 | 1.12 | 1.41 |
| Cajal body organization | 1 | -1.52 | 1.47 | 1.59 | 1.32 | 1.75 | -1.53 | 1.07 |
| quinone cofactor methyltransferase activity | 2 | -1.09 | 1.1 | 1.25 | 1.35 | 1.18 | 1.22 | -1.29 |
| [methionine synthase] reductase activity | 1 | 1.59 | 1.02 | 1.27 | 1.16 | 1.58 | 1.37 | 1.04 |
| pseudocleavage | 8 | 1.05 | 1.14 | 1.52 | 1.06 | 1.52 | 1.39 | 1.23 |
| pseudocleavage involved in syncytial blastoderm formation | 8 | 1.05 | 1.14 | 1.52 | 1.06 | 1.52 | 1.39 | 1.23 |
| U1 snRNA binding | 2 | 1.15 | 1.06 | 1.1 | 1.32 | 1.06 | 1.17 | -1.28 |
| U2 snRNA binding | 1 | 1.41 | -1.11 | 1.3 | 1.34 | 1.31 | 1.54 | -1.14 |
| U4 snRNA binding | 1 | 1.3 | 1.4 | 1.21 | 1.52 | 1.46 | 1.03 | 1.12 |
| regulation of cellular pH | 6 | -1.56 | 1.28 | 1.81 | 1.0 | 1.29 | 1.81 | 1.32 |
| beta-lactam antibiotic metabolic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| beta-lactam antibiotic biosynthetic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| cytoplasmic vesicle membrane | 19 | -1.14 | 1.1 | 1.06 | 1.04 | 1.1 | 1.17 | 1.23 |
| Golgi-associated vesicle membrane | 12 | -1.24 | 1.16 | 1.06 | 1.12 | -1.01 | 1.05 | 1.29 |
| coated vesicle membrane | 19 | -1.14 | 1.1 | 1.06 | 1.04 | 1.1 | 1.17 | 1.23 |
| COPI coated vesicle membrane | 9 | -1.24 | 1.16 | -1.05 | 1.14 | -1.11 | -1.1 | 1.28 |
| clathrin coated vesicle membrane | 9 | -1.07 | 1.0 | 1.1 | -1.06 | 1.25 | 1.39 | 1.09 |
| endocytic vesicle membrane | 1 | -1.65 | 1.56 | 1.35 | 1.0 | 1.12 | 1.4 | 1.32 |
| clathrin-coated endocytic vesicle membrane | 1 | -1.65 | 1.56 | 1.35 | 1.0 | 1.12 | 1.4 | 1.32 |
| synaptic vesicle membrane | 5 | 1.15 | -1.2 | -1.07 | -1.2 | 1.32 | 1.44 | -1.03 |
| axolemma | 1 | -1.08 | -1.3 | -1.04 | 1.04 | 1.56 | 8.15 | 2.89 |
| Rac guanyl-nucleotide exchange factor activity | 1 | 1.08 | -1.14 | -1.07 | -1.55 | -1.02 | 9.19 | -1.47 |
| ribonuclease P complex | 2 | 1.64 | 1.07 | 1.01 | 1.46 | 1.23 | -1.08 | -1.15 |
| multimeric ribonuclease P complex | 1 | 1.84 | 1.03 | 1.1 | 1.42 | 1.17 | 1.05 | -1.14 |
| preribosome | 5 | 1.04 | 1.05 | 1.31 | 1.3 | 1.34 | 1.02 | 1.07 |
| preribosome, small subunit precursor | 1 | 1.41 | 1.31 | 1.41 | 1.25 | 1.47 | 1.04 | 1.34 |
| GTPase regulator activity | 122 | 1.05 | 1.14 | 1.35 | 1.03 | 1.25 | 1.23 | 1.19 |
| chromatin silencing at centromere | 1 | 1.04 | -1.03 | 1.41 | 1.25 | 1.4 | 1.24 | -1.08 |
| eggshell formation | 78 | 1.18 | 1.1 | 1.22 | 1.06 | 1.23 | 1.11 | 1.17 |
| vitelline membrane formation | 12 | 1.14 | 1.01 | -1.05 | -1.02 | -1.08 | 1.01 | -1.0 |
| cytoskeleton-dependent intracellular transport | 28 | -1.03 | 1.2 | 1.26 | 1.11 | 1.22 | 1.23 | 1.11 |
| germarium-derived oocyte differentiation | 23 | 1.09 | 1.17 | 1.19 | 1.1 | 1.16 | 1.23 | 1.19 |
| ovarian follicle cell development | 187 | 1.09 | 1.13 | 1.29 | 1.07 | 1.3 | 1.15 | 1.19 |
| germarium-derived female germ-line cyst encapsulation | 9 | 1.21 | 1.23 | 1.43 | 1.1 | 1.5 | 1.55 | 1.38 |
| border follicle cell delamination | 3 | 1.2 | 1.5 | 1.32 | 1.16 | 1.13 | 2.96 | 1.24 |
| regulation of border follicle cell delamination | 1 | 3.22 | -1.06 | 2.29 | -1.13 | 1.79 | 4.58 | 1.49 |
| ovarian follicle cell stalk formation | 16 | -1.03 | 1.22 | 1.34 | 1.13 | 1.46 | 1.2 | 1.15 |
| anterior/posterior axis specification, follicular epithelium | 1 | 8.32 | -1.09 | 1.22 | 1.02 | 1.27 | 3.95 | 1.61 |
| oocyte fate determination | 22 | 1.1 | 1.16 | 1.25 | 1.08 | 1.23 | 1.33 | 1.24 |
| karyosome formation | 21 | 1.16 | 1.07 | 1.12 | 1.15 | 1.26 | 1.08 | 1.12 |
| germ-line stem cell maintenance | 36 | -1.0 | 1.11 | 1.27 | 1.1 | 1.34 | 1.06 | 1.14 |
| P granule organization | 3 | 2.26 | 1.16 | 2.01 | 1.13 | 1.7 | 2.29 | 1.38 |
| oocyte localization involved in germarium-derived egg chamber formation | 13 | -1.17 | 1.14 | 1.37 | 1.1 | 1.6 | 1.17 | 1.26 |
| spectrosome organization | 3 | -1.62 | 1.17 | 1.61 | 1.01 | 1.24 | 1.17 | 1.31 |
| establishment of oocyte nucleus localization involved in oocyte dorsal/ventral axis specification | 7 | 1.43 | 1.14 | 1.16 | 1.14 | 1.19 | 1.51 | 1.04 |
| ovarian fusome organization | 6 | 1.09 | 1.08 | 1.09 | 1.13 | 1.4 | 1.4 | 1.34 |
| germline ring canal formation | 11 | 1.15 | 1.08 | 1.31 | 1.05 | 1.36 | 1.36 | 1.33 |
| male germline ring canal formation | 1 | 1.93 | 1.13 | 1.83 | -1.03 | 2.06 | 4.41 | 1.79 |
| germarium-derived female germ-line cyst formation | 10 | -1.08 | 1.02 | 1.11 | 1.01 | 1.24 | 1.43 | 1.14 |
| ovulation | 4 | 1.89 | -1.58 | -1.76 | -1.74 | -1.47 | 1.25 | -1.0 |
| sequestering of triglyceride | 5 | 1.07 | 1.21 | 1.1 | 1.17 | 1.11 | 1.16 | 1.09 |
| regulation of cyclic nucleotide metabolic process | 7 | 1.19 | 1.06 | 1.12 | -1.03 | 1.03 | 1.11 | -1.03 |
| negative regulation of cyclic nucleotide metabolic process | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| positive regulation of cyclic nucleotide metabolic process | 6 | 1.16 | 1.07 | 1.14 | -1.06 | 1.03 | 1.1 | -1.03 |
| regulation of cyclic nucleotide biosynthetic process | 7 | 1.19 | 1.06 | 1.12 | -1.03 | 1.03 | 1.11 | -1.03 |
| negative regulation of cyclic nucleotide biosynthetic process | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| positive regulation of cyclic nucleotide biosynthetic process | 6 | 1.16 | 1.07 | 1.14 | -1.06 | 1.03 | 1.1 | -1.03 |
| regulation of nucleotide biosynthetic process | 7 | 1.19 | 1.06 | 1.12 | -1.03 | 1.03 | 1.11 | -1.03 |
| negative regulation of nucleotide biosynthetic process | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| positive regulation of nucleotide biosynthetic process | 6 | 1.16 | 1.07 | 1.14 | -1.06 | 1.03 | 1.1 | -1.03 |
| regulation of nucleotide catabolic process | 43 | 1.08 | 1.16 | 1.36 | 1.06 | 1.3 | 1.31 | 1.21 |
| regulation of cAMP metabolic process | 7 | 1.19 | 1.06 | 1.12 | -1.03 | 1.03 | 1.11 | -1.03 |
| negative regulation of cAMP metabolic process | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| positive regulation of cAMP metabolic process | 6 | 1.16 | 1.07 | 1.14 | -1.06 | 1.03 | 1.1 | -1.03 |
| regulation of cAMP biosynthetic process | 7 | 1.19 | 1.06 | 1.12 | -1.03 | 1.03 | 1.11 | -1.03 |
| negative regulation of cAMP biosynthetic process | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| positive regulation of cAMP biosynthetic process | 6 | 1.16 | 1.07 | 1.14 | -1.06 | 1.03 | 1.1 | -1.03 |
| regulation of actin filament polymerization | 15 | -1.69 | 1.07 | 1.43 | 1.05 | 1.39 | -1.34 | 1.16 |
| regulation of actin filament depolymerization | 2 | -1.38 | -1.03 | 2.18 | 1.01 | 1.59 | -1.39 | -1.04 |
| negative regulation of actin filament depolymerization | 2 | -1.38 | -1.03 | 2.18 | 1.01 | 1.59 | -1.39 | -1.04 |
| negative regulation of actin filament polymerization | 2 | -1.38 | -1.03 | 2.18 | 1.01 | 1.59 | -1.39 | -1.04 |
| positive regulation of actin filament polymerization | 6 | -1.15 | 1.12 | 1.25 | -1.02 | 1.29 | -1.11 | 1.29 |
| inhibition of phospholipase C activity involved in G-protein coupled receptor signaling pathway | 2 | -1.21 | 1.05 | -1.16 | 1.19 | -1.24 | -1.13 | -1.15 |
| epithelial cell differentiation | 29 | 1.27 | 1.07 | 1.39 | 1.01 | 1.31 | 1.36 | 1.32 |
| regulation of epithelial cell differentiation | 1 | 2.56 | -1.26 | -1.08 | -1.5 | 1.05 | 6.9 | -1.01 |
| positive regulation of epithelial cell differentiation | 1 | 2.56 | -1.26 | -1.08 | -1.5 | 1.05 | 6.9 | -1.01 |
| polarized epithelial cell differentiation | 7 | 1.7 | 1.04 | 1.19 | -1.07 | 1.56 | 1.4 | 1.47 |
| cortical cytoskeleton | 9 | -1.53 | 1.16 | 1.66 | 1.07 | 1.5 | 1.1 | 1.12 |
| cortical actin cytoskeleton | 5 | -1.47 | 1.27 | 2.09 | 1.06 | 1.65 | 1.57 | 1.46 |
| cortical cytoskeleton organization | 14 | -1.11 | 1.14 | 1.62 | 1.14 | 1.46 | -1.07 | 1.15 |
| cortical actin cytoskeleton organization | 11 | -1.18 | 1.16 | 1.79 | 1.14 | 1.58 | -1.08 | 1.2 |
| rough endoplasmic reticulum membrane | 12 | -1.63 | 1.34 | -1.12 | 1.02 | -1.3 | -1.73 | 1.28 |
| Mre11 complex | 1 | 1.57 | 1.18 | 2.73 | 1.11 | 1.99 | 2.12 | 1.59 |
| RNA polymerase complex | 23 | 1.32 | 1.14 | 1.23 | 1.25 | 1.26 | 1.12 | 1.07 |
| VCB complex | 1 | 2.69 | 1.18 | 1.72 | 1.15 | -1.01 | -1.18 | -1.08 |
| replisome | 11 | 1.14 | 1.11 | 1.17 | 1.2 | 1.33 | 1.04 | -1.23 |
| actin-dependent ATPase activity | 2 | -1.22 | 1.77 | 3.71 | -1.0 | 2.23 | 2.34 | 2.03 |
| forebrain development | 2 | -1.33 | -1.42 | -1.06 | 1.28 | 1.22 | -1.65 | -2.16 |
| hindbrain development | 2 | -1.31 | 1.27 | 1.45 | 1.03 | 1.47 | -1.24 | 1.48 |
| retromer complex | 2 | 1.62 | 1.07 | 1.77 | 1.2 | 1.41 | 1.97 | 1.23 |
| protein splicing | 1 | -1.72 | -1.51 | 1.07 | 1.74 | 1.86 | -2.25 | -3.98 |
| sheet-forming collagen | 2 | 1.15 | -1.7 | -1.07 | -1.48 | -1.42 | 2.42 | -1.33 |
| endoplasmic reticulum signal peptide binding | 1 | 1.36 | 1.21 | -1.06 | 1.16 | -1.01 | -1.2 | 1.16 |
| establishment or maintenance of microtubule cytoskeleton polarity | 37 | 1.09 | 1.2 | 1.47 | 1.08 | 1.38 | 1.52 | 1.25 |
| establishment or maintenance of cytoskeleton polarity | 37 | 1.09 | 1.2 | 1.47 | 1.08 | 1.38 | 1.52 | 1.25 |
| astral microtubule organization | 2 | 1.07 | 1.11 | 1.15 | 1.21 | 1.27 | 1.05 | 1.05 |
| potassium ion binding | 7 | 1.18 | 1.01 | 1.0 | -1.0 | -1.04 | 1.2 | -1.0 |
| NADH dehydrogenase complex | 37 | 1.15 | 1.04 | 1.27 | 1.07 | 1.11 | 1.72 | -1.29 |
| ER-nuclear sterol response pathway | 1 | 1.18 | 1.17 | 1.43 | 1.18 | 1.36 | -1.03 | 1.42 |
| endoplasmic reticulum unfolded protein response | 1 | -1.05 | 1.27 | -1.17 | -1.24 | -1.49 | -1.01 | 1.19 |
| thiamine pyrophosphate transport | 1 | 1.18 | 1.26 | -1.06 | 1.24 | -1.11 | -1.61 | 1.34 |
| thiamine pyrophosphate binding | 3 | -3.01 | 1.15 | -1.69 | 1.18 | 1.27 | -2.15 | -1.66 |
| cortical microtubule cytoskeleton | 1 | -1.44 | 1.19 | -1.03 | 1.43 | 1.32 | -1.27 | -1.21 |
| mismatched DNA binding | 4 | 1.52 | 1.1 | 1.22 | 1.23 | 1.58 | 1.11 | -1.01 |
| regulation of centriole-centriole cohesion | 1 | 2.67 | 1.08 | 1.26 | 1.12 | 2.08 | 1.82 | 1.58 |
| response to caffeine | 4 | 1.2 | 1.25 | -1.32 | -1.41 | -1.17 | 1.21 | 1.42 |
| filamin binding | 1 | 2.75 | 1.01 | 2.14 | 1.09 | 1.93 | 1.21 | 1.34 |
| ISWI complex | 5 | 1.15 | 1.22 | 1.63 | 1.19 | 1.59 | 1.56 | 1.32 |
| Ino80 complex | 8 | -1.11 | 1.07 | 1.14 | 1.13 | 1.24 | -1.05 | -1.02 |
| extracellular matrix | 65 | 1.16 | -1.04 | 1.03 | -1.11 | 1.03 | 1.19 | -1.0 |
| microtubule organizing center organization | 110 | 1.16 | 1.09 | 1.19 | 1.09 | 1.24 | 1.12 | 1.07 |
| actomyosin structure organization | 47 | 1.76 | -1.18 | 1.14 | -1.18 | 1.17 | 1.52 | 1.04 |
| myosin filament organization | 6 | 1.72 | -1.12 | 1.39 | -1.09 | 1.33 | 1.39 | 1.1 |
| myosin filament assembly | 6 | 1.72 | -1.12 | 1.39 | -1.09 | 1.33 | 1.39 | 1.1 |
| myosin II filament assembly | 1 | -1.92 | 1.09 | 4.17 | -1.09 | 2.38 | -1.63 | 1.77 |
| myosin II filament organization | 1 | -1.92 | 1.09 | 4.17 | -1.09 | 2.38 | -1.63 | 1.77 |
| gene silencing by RNA | 41 | 1.1 | 1.05 | 1.18 | 1.09 | 1.27 | 1.3 | 1.1 |
| chromatin silencing by small RNA | 2 | 1.38 | 1.18 | -1.01 | 1.15 | 1.25 | 2.63 | 1.7 |
| dsRNA fragmentation | 11 | 1.46 | 1.17 | 1.45 | 1.09 | 1.32 | 1.46 | 1.31 |
| primary miRNA processing | 2 | 1.34 | 1.2 | 1.3 | 1.21 | 1.21 | 1.82 | 1.22 |
| pre-miRNA processing | 4 | 1.68 | 1.08 | 1.58 | -1.04 | 1.19 | 1.45 | 1.4 |
| regulation of histone modification | 9 | 1.21 | 1.16 | 1.45 | 1.22 | 1.51 | 1.41 | 1.3 |
| negative regulation of histone modification | 1 | 1.22 | 1.34 | 1.86 | 1.14 | 2.08 | 2.98 | 1.75 |
| positive regulation of histone modification | 4 | 1.11 | 1.17 | 1.59 | 1.27 | 1.57 | 1.16 | 1.28 |
| regulation of histone methylation | 3 | -1.02 | 1.15 | 1.32 | 1.21 | 1.25 | 1.37 | 1.13 |
| positive regulation of histone methylation | 1 | -1.35 | 1.14 | 1.74 | 1.14 | 1.47 | 1.35 | 1.12 |
| regulation of histone deacetylation | 1 | 1.03 | 1.14 | 1.11 | 1.37 | 1.92 | -1.3 | 1.15 |
| positive regulation of histone deacetylation | 1 | 1.03 | 1.14 | 1.11 | 1.37 | 1.92 | -1.3 | 1.15 |
| cysteine desulfurase activity | 1 | 1.7 | 1.2 | 1.13 | 1.08 | -1.16 | -1.08 | 1.29 |
| nuclear pore distribution | 1 | 1.83 | -1.06 | 1.02 | 1.15 | 1.17 | 1.57 | -1.03 |
| BLOC complex | 6 | 1.36 | 1.17 | 1.47 | 1.26 | 1.26 | 1.6 | -1.09 |
| BLOC-1 complex | 6 | 1.36 | 1.17 | 1.47 | 1.26 | 1.26 | 1.6 | -1.09 |
| organelle membrane | 349 | 1.02 | 1.08 | 1.15 | 1.06 | 1.12 | 1.24 | -1.04 |
| stress-activated protein kinase signaling cascade | 29 | 1.02 | 1.15 | 1.45 | 1.1 | 1.49 | 1.28 | 1.37 |
| regeneration | 11 | -1.06 | -1.0 | 1.1 | -1.11 | 1.08 | 1.32 | 1.38 |
| neuron projection regeneration | 3 | -1.12 | 1.14 | 1.65 | 1.16 | 1.31 | -1.01 | 1.22 |
| axon regeneration | 3 | -1.12 | 1.14 | 1.65 | 1.16 | 1.31 | -1.01 | 1.22 |
| septin complex | 6 | -1.44 | 1.09 | 1.11 | 1.19 | 1.4 | -1.24 | 1.03 |
| septin ring organization | 1 | -1.85 | 1.14 | 1.04 | 1.39 | 1.46 | -1.41 | -1.29 |
| microtubule polymerization or depolymerization | 5 | 1.02 | 1.15 | 1.12 | 1.21 | 1.35 | 1.08 | 1.04 |
| regulation of microtubule polymerization or depolymerization | 19 | 1.09 | 1.15 | 1.37 | 1.05 | 1.35 | 1.41 | 1.25 |
| negative regulation of microtubule polymerization or depolymerization | 15 | 1.06 | 1.15 | 1.48 | 1.04 | 1.41 | 1.59 | 1.26 |
| positive regulation of microtubule polymerization or depolymerization | 3 | 1.06 | 1.09 | 1.14 | 1.01 | 1.16 | 1.07 | 1.17 |
| regulation of microtubule polymerization | 2 | 1.39 | -1.03 | 1.05 | -1.01 | 1.01 | -1.16 | 1.04 |
| regulation of microtubule depolymerization | 17 | 1.06 | 1.18 | 1.41 | 1.05 | 1.4 | 1.5 | 1.28 |
| positive regulation of microtubule polymerization | 2 | 1.39 | -1.03 | 1.05 | -1.01 | 1.01 | -1.16 | 1.04 |
| positive regulation of microtubule depolymerization | 1 | -1.6 | 1.37 | 1.36 | 1.04 | 1.54 | 1.63 | 1.47 |
| rRNA pseudouridine synthesis | 2 | 1.38 | 1.09 | 1.14 | 1.35 | 1.29 | -1.02 | -1.04 |
| snRNA pseudouridine synthesis | 2 | -1.23 | 1.23 | 1.84 | 1.26 | 1.56 | -1.27 | 1.05 |
| cytoplasmic microtubule organization | 15 | 1.2 | 1.09 | 1.19 | 1.18 | 1.22 | 1.27 | -1.01 |
| RNA 3'-end processing | 29 | 1.37 | 1.09 | 1.31 | 1.15 | 1.34 | 1.62 | 1.18 |
| mRNA 3'-end processing | 27 | 1.4 | 1.1 | 1.31 | 1.15 | 1.35 | 1.67 | 1.18 |
| sister chromatid biorientation | 1 | 1.13 | 1.05 | -1.04 | 1.47 | 1.34 | -1.18 | -1.11 |
| proteasome localization | 1 | 2.41 | 1.38 | 1.34 | 1.27 | 1.62 | 1.86 | 1.17 |
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | 4 | 1.26 | 1.09 | 1.04 | 1.07 | 1.05 | -1.03 | -1.01 |
| metallo-sulfur cluster assembly | 7 | 1.2 | 1.16 | 1.26 | 1.15 | 1.02 | 1.28 | -1.07 |
| rRNA methylation | 1 | 1.49 | 1.34 | 1.76 | 1.04 | 1.9 | 2.28 | 1.25 |
| neuron projection development | 326 | 1.14 | 1.09 | 1.25 | 1.03 | 1.26 | 1.25 | 1.26 |
| phosphopantetheine binding | 4 | -2.32 | -1.22 | -1.89 | 1.2 | 1.1 | -1.46 | -1.99 |
| peptide modification | 1 | 2.74 | 1.3 | 1.75 | 1.08 | 1.98 | 1.97 | 2.28 |
| POZ domain binding | 1 | 1.78 | 1.16 | 2.67 | 1.12 | 2.53 | 3.19 | 2.15 |
| endoplasmic reticulum palmitoyltransferase complex | 1 | -1.92 | 1.11 | 2.34 | 1.27 | 1.64 | -1.44 | -1.26 |
| RSF complex | 2 | 1.1 | 1.15 | 1.49 | -1.01 | 1.37 | 1.36 | 1.37 |
| auditory behavior | 1 | -1.09 | 1.11 | 1.03 | 1.13 | -1.08 | 1.11 | -1.0 |
| intrinsic to membrane | 1136 | -1.03 | -1.03 | -1.04 | -1.04 | -1.01 | 1.02 | 1.0 |
| intrinsic to plasma membrane | 198 | 1.09 | -1.05 | -1.0 | -1.04 | -1.0 | 1.06 | 1.06 |
| intrinsic to endoplasmic reticulum membrane | 20 | -1.13 | 1.18 | 1.26 | 1.14 | 1.28 | -1.24 | 1.07 |
| intrinsic to Golgi membrane | 5 | 1.35 | 1.04 | -1.19 | 1.04 | 1.29 | 1.43 | 1.52 |
| intrinsic to nuclear inner membrane | 2 | -1.02 | -2.05 | 1.59 | 1.21 | 2.03 | -1.95 | -2.17 |
| intrinsic to peroxisomal membrane | 5 | 1.37 | 1.29 | -1.51 | 1.15 | 1.08 | -1.33 | -1.13 |
| extrinsic to internal side of plasma membrane | 18 | 1.39 | 1.11 | 1.42 | 1.06 | 1.22 | 1.03 | 1.1 |
| external side of cell outer membrane | 1 | 5.43 | -1.29 | -1.04 | -1.06 | 1.21 | 1.62 | 1.41 |
| protein acetyltransferase complex | 1 | 2.72 | 1.09 | 2.08 | 1.56 | 2.05 | 2.2 | 1.21 |
| cell leading edge | 10 | 1.31 | 1.13 | 1.5 | 1.03 | 1.41 | 1.43 | 1.48 |
| cell projection membrane | 4 | -1.03 | 1.05 | 1.14 | 1.11 | 1.34 | 1.84 | 1.38 |
| leading edge membrane | 2 | -1.25 | -1.03 | 1.2 | 1.13 | 1.56 | 3.31 | 1.93 |
| lamellipodium membrane | 1 | -1.44 | 1.23 | 1.5 | 1.23 | 1.55 | 1.34 | 1.29 |
| Ndc80 complex | 7 | 1.37 | 1.01 | 1.12 | 1.26 | 1.43 | 1.16 | -1.13 |
| small GTPase binding | 26 | 1.1 | 1.19 | 1.47 | 1.12 | 1.32 | 1.31 | 1.35 |
| regulation of cyclase activity | 7 | 1.19 | 1.06 | 1.12 | -1.03 | 1.03 | 1.11 | -1.03 |
| negative regulation of cyclase activity | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| positive regulation of cyclase activity | 6 | 1.16 | 1.07 | 1.14 | -1.06 | 1.03 | 1.1 | -1.03 |
| retinal ganglion cell axon guidance | 11 | 1.15 | 1.11 | 1.28 | 1.05 | 1.21 | 1.18 | 1.12 |
| intrinsic to organelle membrane | 38 | 1.02 | 1.11 | 1.12 | 1.13 | 1.26 | -1.1 | 1.02 |
| integral to organelle membrane | 28 | -1.02 | 1.06 | 1.03 | 1.14 | 1.16 | -1.18 | -1.09 |
| intrinsic to mitochondrial inner membrane | 1 | -1.27 | 1.34 | 1.46 | 1.01 | -1.06 | 2.1 | -1.33 |
| integral to mitochondrial inner membrane | 1 | -1.27 | 1.34 | 1.46 | 1.01 | -1.06 | 2.1 | -1.33 |
| intrinsic to mitochondrial outer membrane | 1 | 1.09 | 1.16 | 1.56 | 1.4 | 1.39 | 1.34 | -1.11 |
| integral to mitochondrial outer membrane | 1 | 1.09 | 1.16 | 1.56 | 1.4 | 1.39 | 1.34 | -1.11 |
| regulation of cellular metabolic process | 989 | 1.17 | 1.09 | 1.19 | 1.06 | 1.18 | 1.19 | 1.14 |
| negative regulation of cellular metabolic process | 253 | 1.16 | 1.07 | 1.18 | 1.05 | 1.21 | 1.19 | 1.14 |
| positive regulation of cellular metabolic process | 273 | 1.17 | 1.04 | 1.14 | 1.02 | 1.14 | 1.21 | 1.18 |
| regulation of cellular biosynthetic process | 770 | 1.16 | 1.07 | 1.16 | 1.05 | 1.17 | 1.16 | 1.13 |
| negative regulation of cellular biosynthetic process | 219 | 1.16 | 1.07 | 1.18 | 1.04 | 1.23 | 1.2 | 1.14 |
| positive regulation of cellular biosynthetic process | 251 | 1.16 | 1.03 | 1.12 | 1.02 | 1.13 | 1.2 | 1.17 |
| regulation of cellular catabolic process | 68 | 1.17 | 1.13 | 1.25 | 1.07 | 1.22 | 1.25 | 1.16 |
| negative regulation of cellular catabolic process | 7 | -1.14 | 1.23 | -1.01 | 1.15 | -1.01 | -1.46 | 1.01 |
| positive regulation of cellular catabolic process | 7 | 1.19 | 1.14 | 1.23 | 1.17 | 1.1 | 1.13 | 1.03 |
| RNAi effector complex | 7 | -1.01 | 1.19 | 1.52 | 1.03 | 1.34 | 1.15 | 1.36 |
| negative regulation of protein complex assembly | 2 | -1.38 | -1.03 | 2.18 | 1.01 | 1.59 | -1.39 | -1.04 |
| positive regulation of protein complex assembly | 11 | 1.26 | 1.07 | 1.19 | -1.02 | 1.17 | 1.16 | 1.39 |
| regulation of vesicle fusion | 1 | 1.15 | 1.03 | 1.12 | 1.08 | 1.15 | 1.15 | -1.08 |
| negative regulation of vesicle fusion | 1 | 1.15 | 1.03 | 1.12 | 1.08 | 1.15 | 1.15 | -1.08 |
| regulation of cell projection organization | 59 | 1.08 | 1.13 | 1.42 | 1.08 | 1.39 | 1.17 | 1.18 |
| negative regulation of cell projection organization | 13 | -1.04 | 1.17 | 1.5 | 1.06 | 1.4 | 1.02 | 1.24 |
| positive regulation of cell projection organization | 6 | 1.15 | 1.11 | 1.56 | 1.11 | 1.48 | -1.16 | 1.25 |
| regulation of defense response | 35 | 1.89 | -1.19 | -1.0 | -1.08 | -1.03 | 1.26 | 1.14 |
| negative regulation of defense response | 5 | 2.13 | -1.29 | 1.2 | -1.05 | -1.19 | -1.21 | -1.11 |
| positive regulation of defense response | 6 | 2.51 | -1.09 | 1.1 | -1.06 | 1.06 | 1.76 | 1.22 |
| N-terminal protein amino acid modification | 3 | -1.27 | 1.36 | 1.99 | 1.28 | 1.6 | -1.05 | -1.03 |
| translation initiation factor binding | 10 | 1.25 | 1.1 | 1.24 | 1.01 | 1.08 | 1.15 | 1.37 |
| eukaryotic initiation factor 4G binding | 7 | 1.14 | 1.02 | 1.05 | -1.04 | -1.09 | -1.04 | 1.26 |
| bursicon neuropeptide hormone complex | 2 | -1.74 | -1.0 | -1.2 | 1.19 | 1.42 | -1.55 | -1.19 |
| regulation of protein ubiquitination | 10 | 1.62 | 1.07 | 1.68 | 1.03 | 1.28 | 1.35 | 1.26 |
| negative regulation of protein ubiquitination | 2 | 1.45 | 1.11 | 1.45 | 1.15 | 1.24 | 1.19 | 1.21 |
| positive regulation of protein ubiquitination | 2 | -1.0 | -1.14 | 1.48 | -1.16 | 1.16 | -1.27 | -1.27 |
| regulation of protein modification process | 92 | 1.25 | 1.15 | 1.39 | 1.12 | 1.32 | 1.39 | 1.2 |
| negative regulation of protein modification process | 10 | 1.3 | 1.16 | 1.35 | 1.1 | 1.31 | 1.4 | 1.13 |
| positive regulation of protein modification process | 11 | 1.08 | 1.11 | 1.65 | 1.11 | 1.39 | 1.35 | 1.2 |
| carboxylic acid binding | 52 | -1.23 | -1.12 | -1.35 | 1.02 | 1.06 | -1.11 | -1.28 |
| cytoplasmic vesicle | 114 | 1.02 | 1.01 | 1.1 | 1.05 | 1.12 | 1.08 | 1.1 |
| L-ascorbic acid binding | 26 | -1.0 | 1.01 | -1.07 | -1.0 | -1.07 | 1.07 | -1.07 |
| cobalamin binding | 1 | -1.04 | 1.01 | 1.04 | 1.07 | -1.1 | -1.21 | 1.08 |
| alkali metal ion binding | 7 | 1.18 | 1.01 | 1.0 | -1.0 | -1.04 | 1.2 | -1.0 |
| response to methotrexate | 11 | -1.41 | 1.08 | -1.36 | -1.03 | 1.36 | -1.33 | 1.53 |
| box H/ACA snoRNP complex | 1 | 1.3 | 1.21 | 1.13 | 1.29 | 1.21 | 1.06 | 1.05 |
| M band | 6 | 1.68 | 1.03 | -1.27 | -1.02 | 1.18 | 1.68 | 1.16 |
| mitogen-activated protein kinase kinase binding | 1 | 1.22 | 1.33 | 2.18 | 1.07 | 1.72 | 2.05 | 1.61 |
| regulation of mRNA 3'-end processing | 5 | 1.3 | 1.09 | 1.27 | 1.06 | 1.11 | 1.44 | 1.1 |
| negative regulation of mRNA 3'-end processing | 1 | 1.92 | 1.01 | 1.57 | -1.13 | 1.41 | 2.91 | 1.31 |
| positive regulation of mRNA 3'-end processing | 4 | 1.18 | 1.11 | 1.21 | 1.11 | 1.04 | 1.21 | 1.05 |
| regulation of heterochromatin assembly | 3 | 1.21 | 1.11 | 1.43 | 1.11 | 1.52 | 1.35 | 1.11 |
| negative regulation of heterochromatin assembly | 1 | 1.22 | 1.33 | 2.18 | 1.07 | 1.72 | 2.05 | 1.61 |
| positive regulation of heterochromatin assembly | 2 | 1.21 | 1.02 | 1.16 | 1.13 | 1.43 | 1.09 | -1.08 |
| cullin-RING ubiquitin ligase complex | 30 | 1.33 | 1.16 | 1.26 | 1.17 | 1.29 | 1.15 | 1.13 |
| Cul3-RING ubiquitin ligase complex | 3 | 1.11 | 1.17 | 1.23 | 1.18 | 1.27 | 1.3 | 1.21 |
| nuclear envelope reassembly | 4 | 1.3 | 1.11 | 1.08 | 1.3 | 1.24 | -1.17 | -1.07 |
| myosin III binding | 1 | -1.23 | 1.03 | 1.03 | -1.15 | -1.01 | -1.11 | -1.09 |
| myosin VI complex | 3 | -1.98 | 1.02 | 1.13 | 1.07 | 1.24 | -1.12 | -1.15 |
| chromatin DNA binding | 10 | 1.28 | 1.12 | 1.17 | 1.21 | 1.23 | 1.07 | 1.13 |
| nucleosome binding | 2 | -1.08 | 1.25 | 1.65 | 1.07 | 1.57 | 1.08 | 1.51 |
| nucleosomal DNA binding | 1 | -1.26 | 1.34 | 1.15 | 1.09 | 1.13 | -1.62 | 1.24 |
| chromatin assembly | 32 | 1.01 | 1.07 | 1.31 | 1.14 | 1.26 | -1.03 | 1.08 |
| protein complex localization | 2 | 2.1 | 1.14 | 1.17 | 1.2 | 1.38 | 1.71 | 1.06 |
| heterochromatin assembly | 4 | -1.26 | 1.27 | 1.36 | 1.02 | 1.42 | 1.07 | 1.46 |
| SUMO activating enzyme complex | 2 | 1.33 | 1.09 | 1.29 | 1.43 | 1.67 | 1.2 | -1.13 |
| nonmotile primary cilium | 2 | -1.07 | -1.03 | -1.02 | 1.13 | -1.1 | -1.09 | -1.15 |
| PcG protein complex | 15 | 1.41 | 1.18 | 1.36 | 1.09 | 1.31 | 1.34 | 1.26 |
| Myb complex | 11 | 1.43 | 1.19 | 1.32 | 1.2 | 1.4 | 1.5 | 1.18 |
| microvillus membrane | 1 | 1.23 | 1.27 | 1.15 | 1.19 | 1.2 | 1.15 | 1.07 |
| actin cytoskeleton reorganization | 9 | 1.38 | 1.0 | 1.56 | 1.0 | 1.39 | 1.59 | 1.34 |
| minus-end directed microtubule sliding | 1 | -1.34 | -1.01 | 1.11 | 1.33 | 1.45 | -1.15 | -1.3 |
| plus-end directed microtubule sliding | 1 | 1.34 | 1.07 | 1.06 | 1.01 | 1.45 | -1.14 | -1.53 |
| positive regulation of exit from mitosis | 1 | 2.76 | -2.65 | -2.69 | -2.98 | -1.9 | 34.44 | -1.85 |
| peptidyl-proline dioxygenase activity | 27 | 1.0 | -1.0 | -1.07 | 1.0 | -1.1 | -1.02 | -1.07 |
| peptidyl-proline 4-dioxygenase activity | 27 | 1.0 | -1.0 | -1.07 | 1.0 | -1.1 | -1.02 | -1.07 |
| contractile ring maintenance involved in cell cycle cytokinesis | 1 | -1.85 | 1.14 | 1.04 | 1.39 | 1.46 | -1.41 | -1.29 |
| DNA integrity checkpoint | 70 | 1.22 | 1.15 | 1.38 | 1.19 | 1.41 | 1.3 | 1.15 |
| mitotic cell cycle G1/S transition DNA damage checkpoint | 2 | 1.09 | 1.49 | 1.71 | 1.02 | 1.44 | 1.9 | 1.73 |
| G2/M transition DNA damage checkpoint | 63 | 1.23 | 1.15 | 1.36 | 1.2 | 1.39 | 1.27 | 1.14 |
| mitotic cell cycle G1/S transition checkpoint | 2 | 1.09 | 1.49 | 1.71 | 1.02 | 1.44 | 1.9 | 1.73 |
| G2/M transition checkpoint | 63 | 1.23 | 1.15 | 1.36 | 1.2 | 1.39 | 1.27 | 1.14 |
| spindle checkpoint | 10 | 1.07 | 1.09 | 1.16 | 1.15 | 1.32 | 1.1 | 1.03 |
| AMP-activated protein kinase complex | 1 | 1.2 | 1.33 | 1.18 | -1.01 | -1.15 | 1.3 | 1.08 |
| cell-substrate adhesion | 18 | -1.4 | 1.01 | 1.22 | -1.05 | 1.29 | -1.09 | 1.19 |
| neuromuscular junction | 29 | 1.64 | -1.02 | 1.13 | -1.17 | 1.09 | 1.63 | 1.22 |
| spindle pole centrosome | 3 | 1.78 | 1.17 | 1.45 | -1.07 | 1.38 | 1.95 | 1.49 |
| NMS complex | 3 | 1.88 | 1.01 | 1.19 | 1.2 | 1.44 | 1.48 | -1.09 |
| nuclear centromeric heterochromatin | 1 | -1.2 | 1.03 | 1.07 | -1.11 | 1.52 | -1.28 | -1.06 |
| receptor internalization | 3 | 1.27 | -1.02 | 1.22 | -1.05 | 1.11 | 1.66 | 1.17 |
| ubiquitin conjugating enzyme binding | 1 | 1.42 | -1.23 | 1.7 | 1.22 | 1.08 | 1.57 | -1.21 |
| ubiquitin protein ligase binding | 9 | 1.11 | 1.17 | 1.66 | 1.19 | 1.63 | 1.3 | 1.36 |
| synaptic vesicle fusion to presynaptic membrane | 2 | -3.55 | -1.94 | -2.8 | 1.67 | -1.37 | -8.1 | -3.92 |
| regulation of synaptic vesicle fusion to presynaptic membrane | 1 | 1.15 | 1.03 | 1.12 | 1.08 | 1.15 | 1.15 | -1.08 |
| negative regulation of synaptic vesicle fusion to presynaptic membrane | 1 | 1.15 | 1.03 | 1.12 | 1.08 | 1.15 | 1.15 | -1.08 |
| regulation of neuronal synaptic plasticity in response to neurotrophin | 1 | 2.75 | -1.14 | -1.16 | -1.07 | -1.18 | 1.11 | 1.67 |
| zymogen activation | 4 | 3.67 | -1.44 | -1.39 | -1.42 | -1.27 | 2.44 | 1.48 |
| regulation of neurological system process | 37 | 1.13 | 1.11 | 1.3 | 1.03 | 1.17 | 1.29 | 1.22 |
| negative regulation of neurological system process | 2 | -1.06 | 1.07 | 1.19 | 1.01 | 1.07 | 1.19 | 1.07 |
| positive regulation of neurological system process | 2 | -1.81 | 1.29 | 2.08 | -1.04 | 1.39 | 1.52 | 1.29 |
| regulation of protein stability | 27 | 1.47 | 1.17 | 1.27 | 1.1 | 1.25 | 1.18 | 1.11 |
| regulation of cyclin-dependent protein kinase activity involved in G2/M | 1 | 1.18 | 1.23 | 1.07 | 1.21 | 1.0 | 1.04 | -1.03 |
| response to nutrient levels | 42 | 1.1 | -1.0 | 1.12 | -1.03 | 1.15 | 1.24 | 1.18 |
| cellular response to extracellular stimulus | 16 | 1.0 | 1.13 | 1.15 | 1.05 | 1.11 | 1.4 | 1.14 |
| cellular response to nutrient levels | 15 | -1.0 | 1.14 | 1.14 | 1.06 | 1.09 | 1.4 | 1.13 |
| cellular response to nutrient | 1 | -36.3 | 1.49 | 1.05 | 1.2 | 1.58 | -8.6 | -1.57 |
| I band | 19 | 1.85 | -1.41 | -1.18 | -1.11 | 1.31 | 1.7 | -1.14 |
| lipid particle transport along microtubule | 3 | -1.08 | 1.32 | -1.03 | -1.05 | 1.05 | -1.42 | -1.07 |
| late endosome membrane | 1 | 1.78 | 1.14 | -1.44 | 1.08 | 1.0 | 3.89 | 1.83 |
| microbody membrane | 14 | 1.87 | 1.14 | -1.32 | 1.28 | 1.36 | -1.33 | -1.09 |
| telomeric heterochromatin | 2 | -1.21 | 1.23 | 1.23 | 1.13 | 1.5 | -1.12 | 1.13 |
| regulation of chromatin silencing | 19 | 1.34 | 1.17 | 1.13 | 1.09 | 1.19 | 1.43 | 1.42 |
| negative regulation of chromatin silencing | 6 | 1.15 | 1.16 | 1.19 | 1.25 | 1.26 | 1.19 | 1.22 |
| positive regulation of chromatin silencing | 3 | 1.21 | 1.14 | -1.1 | -1.08 | 1.08 | 1.29 | 1.99 |
| filamentous actin | 6 | -1.28 | -1.05 | 1.22 | -1.12 | 1.0 | 1.16 | 1.38 |
| beta-alanyl-histamine hydrolase activity | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| nuclear membrane | 16 | 1.66 | -1.06 | 1.3 | 1.05 | 1.4 | 1.42 | 1.19 |
| mitochondrial membrane | 181 | 1.05 | 1.06 | 1.17 | 1.06 | 1.11 | 1.4 | -1.2 |
| organelle envelope | 277 | 1.12 | 1.07 | 1.19 | 1.07 | 1.16 | 1.37 | -1.11 |
| organelle outer membrane | 24 | 1.17 | -1.02 | 1.15 | 1.1 | 1.19 | 1.35 | 1.0 |
| organelle envelope lumen | 13 | 1.07 | 1.12 | 1.22 | 1.17 | 1.15 | 1.4 | -1.16 |
| membrane-enclosed lumen | 601 | 1.19 | 1.12 | 1.23 | 1.16 | 1.24 | 1.22 | -1.0 |
| envelope | 288 | 1.15 | 1.07 | 1.18 | 1.06 | 1.15 | 1.36 | -1.1 |
| nuclear lumen | 436 | 1.23 | 1.14 | 1.25 | 1.18 | 1.27 | 1.15 | 1.08 |
| vesicle | 126 | -1.01 | 1.01 | 1.1 | 1.06 | 1.15 | 1.08 | 1.1 |
| organelle subcompartment | 9 | 1.35 | -1.0 | 1.2 | 1.12 | 1.08 | 1.27 | 1.25 |
| Golgi cisterna | 6 | 1.05 | 1.2 | 1.02 | 1.18 | 1.21 | 1.34 | 1.32 |
| locomotion involved in locomotory behavior | 14 | -1.12 | 1.15 | 1.42 | 1.11 | 1.53 | 1.07 | 1.2 |
| membrane-bounded vesicle | 107 | 1.03 | 1.05 | 1.13 | 1.04 | 1.17 | 1.14 | 1.16 |
| bombesin receptor signaling pathway | 2 | 1.03 | 1.12 | 1.37 | -1.03 | -1.15 | 1.42 | 1.13 |
| mRNA export from nucleus in response to heat stress | 2 | -1.27 | 1.03 | 1.6 | -1.01 | 1.13 | 1.81 | 1.1 |
| regulation of TOR signaling cascade | 3 | -2.06 | 1.16 | 1.27 | 1.19 | 1.19 | -1.72 | 1.0 |
| negative regulation of TOR signaling cascade | 1 | 1.13 | 1.46 | 1.84 | 1.15 | 1.31 | 1.41 | 1.45 |
| positive regulation of TOR signaling cascade | 2 | -3.14 | 1.04 | 1.05 | 1.2 | 1.13 | -2.68 | -1.2 |
| regulation of ARF protein signal transduction | 14 | -1.12 | 1.16 | 1.23 | 1.01 | 1.21 | 1.19 | 1.55 |
| NELF complex | 4 | 1.21 | 1.13 | 1.22 | 1.16 | 1.17 | 1.14 | 1.08 |
| myosin light chain binding | 13 | -1.01 | 1.07 | 1.48 | -1.0 | 1.2 | 1.25 | 1.17 |
| myosin head/neck binding | 2 | -1.4 | -1.01 | 1.0 | 1.02 | 1.17 | -1.17 | -1.2 |
| myosin II light chain binding | 1 | -1.26 | 1.24 | 1.29 | 1.08 | 1.1 | -1.05 | 1.16 |
| myosin heavy chain binding | 5 | -1.16 | 1.05 | 1.2 | 1.06 | 1.23 | -1.26 | -1.0 |
| myosin II heavy chain binding | 1 | -1.26 | 1.24 | 1.29 | 1.08 | 1.1 | -1.05 | 1.16 |
| small-subunit processome | 4 | -1.04 | -1.0 | 1.29 | 1.31 | 1.31 | 1.02 | 1.02 |
| mitochondrial DNA metabolic process | 5 | -1.01 | 1.2 | 1.25 | 1.15 | 1.21 | 1.2 | -1.15 |
| cardiolipin metabolic process | 1 | -3.36 | 1.4 | 1.32 | 1.25 | 1.35 | -2.25 | 1.21 |
| cardiolipin biosynthetic process | 1 | -3.36 | 1.4 | 1.32 | 1.25 | 1.35 | -2.25 | 1.21 |
| clathrin heavy chain binding | 2 | -1.49 | 1.38 | 1.52 | 1.18 | 1.43 | -1.03 | 1.15 |
| microtubule basal body organization | 2 | 1.48 | 1.06 | 1.18 | 1.02 | 1.1 | 1.35 | 1.01 |
| cortical protein anchoring | 1 | -1.33 | 1.57 | 2.09 | 1.17 | 1.77 | 1.11 | 1.58 |
| regulation of nuclease activity | 1 | 1.58 | 1.16 | -1.09 | 1.12 | 1.04 | 1.36 | -1.13 |
| negative regulation of nuclease activity | 1 | 1.58 | 1.16 | -1.09 | 1.12 | 1.04 | 1.36 | -1.13 |
| SAM domain binding | 1 | 1.45 | 1.28 | 1.83 | 1.25 | 2.05 | 1.81 | 1.62 |
| regulation of response to food | 2 | -2.4 | -1.4 | 3.96 | -1.44 | -2.81 | -4.01 | -2.19 |
| regulation of response to external stimulus | 9 | -1.23 | 1.03 | 1.62 | -1.03 | -1.11 | -1.21 | -1.18 |
| negative regulation of response to external stimulus | 2 | 1.19 | 1.12 | 1.18 | 1.18 | 1.31 | 1.07 | -1.1 |
| regulation of response to extracellular stimulus | 4 | -1.71 | -1.02 | 2.0 | -1.11 | -1.48 | -1.65 | -1.49 |
| negative regulation of response to extracellular stimulus | 1 | -1.06 | 1.16 | 1.2 | 1.34 | 1.78 | -1.33 | -1.11 |
| regulation of response to nutrient levels | 4 | -1.71 | -1.02 | 2.0 | -1.11 | -1.48 | -1.65 | -1.49 |
| negative regulation of response to nutrient levels | 1 | -1.06 | 1.16 | 1.2 | 1.34 | 1.78 | -1.33 | -1.11 |
| chromosome passenger complex | 2 | -1.25 | -1.08 | -1.11 | 1.09 | 1.18 | -1.16 | -1.19 |
| activation of protein kinase activity | 19 | 1.27 | 1.12 | 1.2 | 1.07 | 1.14 | 1.38 | 1.17 |
| cell division site | 12 | -1.06 | 1.09 | 1.54 | 1.06 | 1.53 | 1.28 | 1.13 |
| cell division site part | 12 | -1.06 | 1.09 | 1.54 | 1.06 | 1.53 | 1.28 | 1.13 |
| septin cytoskeleton | 6 | -1.44 | 1.09 | 1.11 | 1.19 | 1.4 | -1.24 | 1.03 |
| small conjugating protein binding | 3 | 1.59 | 1.33 | 1.88 | 1.29 | 1.84 | 1.63 | 1.43 |
| SUMO binding | 1 | 1.3 | 1.31 | 1.72 | 1.5 | 1.61 | 1.9 | 1.2 |
| septin cytoskeleton organization | 1 | -1.85 | 1.14 | 1.04 | 1.39 | 1.46 | -1.41 | -1.29 |
| contractile ring localization involved in cell cycle cytokinesis | 2 | -1.56 | 1.16 | 1.2 | 1.2 | 1.28 | -1.0 | 1.03 |
| maintenance of contractile ring localization involved in cell cycle cytokinesis | 1 | -1.85 | 1.14 | 1.04 | 1.39 | 1.46 | -1.41 | -1.29 |
| transposition | 1 | 1.07 | -1.04 | -1.16 | 1.02 | 1.48 | -1.27 | -1.22 |
| telomere organization | 14 | 1.19 | 1.12 | 1.39 | 1.17 | 1.35 | 1.2 | 1.11 |
| regulation of synaptic transmission, cholinergic | 1 | -1.1 | 1.17 | -1.13 | -1.04 | -1.08 | -1.08 | 1.11 |
| regulation of actin filament bundle assembly | 3 | 1.54 | 1.04 | 1.3 | -1.03 | 2.35 | 1.13 | 1.3 |
| regulation of calcium ion transport via store-operated calcium channel activity | 3 | 1.93 | -1.03 | 1.33 | 1.03 | 1.24 | 2.17 | 1.55 |
| positive regulation of calcium ion transport via store-operated calcium channel activity | 2 | 2.64 | -1.06 | 1.57 | 1.01 | 1.45 | 3.25 | 1.94 |
| activation of store-operated calcium channel activity | 1 | 2.63 | 1.01 | 1.53 | -1.05 | 1.46 | 2.52 | 1.76 |
| regulation of nucleobase-containing compound transport | 1 | 1.52 | 1.17 | 1.71 | 1.25 | 2.2 | 8.74 | 2.48 |
| negative regulation of nucleobase-containing compound transport | 1 | 1.52 | 1.17 | 1.71 | 1.25 | 2.2 | 8.74 | 2.48 |
| methylation | 53 | 1.32 | 1.18 | 1.3 | 1.18 | 1.32 | 1.25 | 1.12 |
| regulation of cellular protein metabolic process | 160 | 1.3 | 1.11 | 1.31 | 1.08 | 1.29 | 1.35 | 1.22 |
| negative regulation of cellular protein metabolic process | 47 | 1.33 | 1.04 | 1.25 | 1.02 | 1.25 | 1.33 | 1.24 |
| positive regulation of cellular protein metabolic process | 25 | 1.12 | 1.15 | 1.46 | 1.08 | 1.34 | 1.24 | 1.28 |
| regulation of protein polymerization | 17 | -1.53 | 1.06 | 1.38 | 1.05 | 1.34 | -1.31 | 1.14 |
| negative regulation of protein polymerization | 2 | -1.38 | -1.03 | 2.18 | 1.01 | 1.59 | -1.39 | -1.04 |
| positive regulation of protein polymerization | 8 | -1.02 | 1.08 | 1.2 | -1.02 | 1.21 | -1.12 | 1.22 |
| double-stranded RNA-specific ribonuclease activity | 1 | 6.73 | -1.62 | -1.13 | -2.09 | 1.01 | 8.39 | 1.39 |
| mismatch repair complex | 1 | 2.58 | -1.1 | 1.19 | 1.19 | 2.02 | -1.05 | -1.35 |
| MutSalpha complex | 1 | 2.58 | -1.1 | 1.19 | 1.19 | 2.02 | -1.05 | -1.35 |
| MutSbeta complex | 1 | 2.58 | -1.1 | 1.19 | 1.19 | 2.02 | -1.05 | -1.35 |
| regulation of ARF GTPase activity | 8 | -1.06 | 1.12 | 1.2 | 1.04 | 1.17 | 1.01 | 1.53 |
| regulation of Rab GTPase activity | 25 | 1.14 | 1.15 | 1.4 | 1.07 | 1.25 | 1.3 | 1.16 |
| regulation of Rac GTPase activity | 1 | 1.08 | -1.14 | -1.07 | -1.55 | -1.02 | 9.19 | -1.47 |
| regulation of Rap GTPase activity | 1 | -1.22 | 2.43 | -1.06 | -1.67 | 1.03 | 1.44 | -1.08 |
| regulation of Ras GTPase activity | 38 | 1.08 | 1.17 | 1.37 | 1.04 | 1.27 | 1.32 | 1.23 |
| regulation of Rho GTPase activity | 4 | 1.14 | 1.13 | 1.64 | 1.03 | 1.74 | 2.44 | 1.19 |
| positive regulation of Ras GTPase activity | 3 | 1.01 | 1.31 | 1.81 | 1.2 | 1.9 | 1.82 | 1.38 |
| positive regulation of Rho GTPase activity | 2 | -1.11 | 1.35 | 2.32 | 1.27 | 2.42 | 1.41 | 1.46 |
| molybdopterin cofactor biosynthetic process | 5 | 1.14 | 1.31 | 1.37 | 1.14 | 1.16 | -1.34 | 1.2 |
| regulation of hormone metabolic process | 4 | -4.19 | -1.34 | 1.3 | 1.27 | 1.23 | -3.79 | -2.08 |
| negative regulation of hormone metabolic process | 2 | -15.63 | -2.03 | 1.53 | 1.8 | 1.58 | -15.3 | -4.44 |
| negative regulation of hormone biosynthetic process | 1 | -84.21 | -3.59 | 1.42 | 2.07 | 1.14 | -58.36 | -15.07 |
| intracellular lipid transport | 1 | -2.39 | 1.47 | 1.51 | 1.07 | -1.25 | -2.16 | 1.24 |
| regulation of lipid transport | 1 | 2.58 | -1.39 | 1.67 | -1.07 | 1.62 | 11.34 | 1.83 |
| regulation of sterol transport | 1 | 2.58 | -1.39 | 1.67 | -1.07 | 1.62 | 11.34 | 1.83 |
| regulation of cholesterol transport | 1 | 2.58 | -1.39 | 1.67 | -1.07 | 1.62 | 11.34 | 1.83 |
| regulation of intracellular transport | 36 | 1.16 | 1.11 | 1.23 | 1.1 | 1.23 | 1.22 | 1.22 |
| negative regulation of intracellular transport | 6 | 2.03 | 1.13 | 1.29 | 1.08 | 1.28 | 1.86 | 1.59 |
| positive regulation of intracellular transport | 23 | 1.1 | 1.07 | 1.2 | 1.09 | 1.25 | 1.08 | 1.14 |
| DNA geometric change | 3 | 1.03 | 1.28 | -1.01 | 1.04 | -1.04 | 1.08 | 1.21 |
| protein complex binding | 19 | 1.04 | 1.06 | 1.22 | 1.11 | 1.09 | 1.09 | 1.1 |
| regulation of transporter activity | 7 | 1.44 | -1.04 | 1.16 | -1.02 | 1.15 | 1.52 | 1.13 |
| positive regulation of transporter activity | 2 | 2.64 | -1.06 | 1.57 | 1.01 | 1.45 | 3.25 | 1.94 |
| regulation of ion transmembrane transporter activity | 7 | 1.44 | -1.04 | 1.16 | -1.02 | 1.15 | 1.52 | 1.13 |
| positive regulation of ion transmembrane transporter activity | 2 | 2.64 | -1.06 | 1.57 | 1.01 | 1.45 | 3.25 | 1.94 |
| beta-N-acetylgalactosaminidase activity | 1 | 3.13 | 1.28 | 1.73 | 1.05 | 1.33 | 2.12 | 2.37 |
| actin filament bundle | 1 | 1.76 | 1.07 | 1.13 | 1.08 | 1.49 | 1.28 | 1.15 |
| filopodium tip | 2 | 1.13 | -1.0 | 1.96 | -1.15 | 1.62 | 1.33 | 1.8 |
| regulation of proteasomal ubiquitin-dependent protein catabolic process | 4 | 1.63 | 1.06 | 1.19 | 1.23 | 1.22 | 1.17 | 1.04 |
| negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 3 | 1.42 | 1.07 | 1.19 | 1.26 | 1.18 | -1.01 | -1.02 |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 2.44 | 1.03 | 1.21 | 1.16 | 1.33 | 1.95 | 1.23 |
| melanosome organization | 1 | -1.21 | 1.04 | -1.11 | 1.04 | -1.07 | -1.17 | -1.01 |
| protein modification by small protein conjugation | 47 | 1.14 | 1.13 | 1.39 | 1.16 | 1.27 | 1.17 | 1.12 |
| demethylase activity | 6 | 1.22 | 1.23 | 1.76 | 1.12 | 1.66 | 1.8 | 1.31 |
| histone demethylase activity | 6 | 1.22 | 1.23 | 1.76 | 1.12 | 1.66 | 1.8 | 1.31 |
| histone demethylase activity (H3-K4 specific) | 2 | 1.08 | 1.42 | 1.76 | 1.01 | 1.58 | 2.53 | 1.88 |
| histone demethylase activity (H3-K9 specific) | 2 | 1.08 | 1.24 | 1.45 | 1.25 | 1.55 | 1.06 | 1.05 |
| endocytic recycling | 5 | 1.33 | 1.24 | 1.42 | 1.12 | 1.35 | 1.65 | 1.25 |
| regulation of cytokinesis | 1 | -1.32 | 1.18 | 1.39 | 1.03 | 1.12 | 1.4 | 1.35 |
| endoplasmic reticulum calcium ion homeostasis | 1 | 4.45 | 1.05 | -1.49 | 1.14 | 1.22 | 1.67 | 1.4 |
| regulation of Rab protein signal transduction | 25 | 1.14 | 1.15 | 1.4 | 1.07 | 1.25 | 1.3 | 1.16 |
| regulation of Rap protein signal transduction | 1 | -1.22 | 2.43 | -1.06 | -1.67 | 1.03 | 1.44 | -1.08 |
| response to peptidoglycan | 1 | 9.16 | -1.23 | -1.57 | -1.54 | -1.3 | 2.66 | -1.01 |
| response to lipopolysaccharide | 2 | 1.04 | 1.17 | 1.37 | 1.17 | 1.75 | 1.28 | 1.1 |
| muramyl dipeptide binding | 1 | 9.16 | -1.23 | -1.57 | -1.54 | -1.3 | 2.66 | -1.01 |
| multicellular organismal process | 2813 | 1.13 | 1.04 | 1.13 | 1.03 | 1.14 | 1.16 | 1.1 |
| developmental process | 2455 | 1.15 | 1.04 | 1.14 | 1.03 | 1.17 | 1.2 | 1.12 |
| multicellular organism reproduction | 694 | 1.11 | 1.06 | 1.17 | 1.03 | 1.19 | 1.16 | 1.16 |
| cytokinetic process | 22 | 1.13 | -1.06 | 1.1 | -1.09 | 1.19 | 1.19 | 1.11 |
| maintenance of protein location in cell | 37 | 1.15 | 1.12 | 1.29 | 1.08 | 1.31 | 1.23 | 1.24 |
| DNA duplex unwinding | 3 | 1.03 | 1.28 | -1.01 | 1.04 | -1.04 | 1.08 | 1.21 |
| endosome transport via multivesicular body sorting pathway | 3 | 1.32 | -1.02 | 1.33 | 1.3 | 1.34 | 1.03 | -1.24 |
| protein exit from endoplasmic reticulum | 1 | -1.49 | 1.52 | 1.25 | 1.12 | 1.19 | 1.46 | 1.78 |
| microvillus organization | 4 | -2.72 | -1.1 | -1.32 | -1.18 | -1.23 | -2.04 | 1.51 |
| follicle cell microvillus organization | 2 | 1.82 | -1.12 | -1.27 | -1.14 | -1.29 | -1.07 | 1.07 |
| regulation of microvillus organization | 1 | 3.92 | -1.13 | -1.16 | -1.25 | -1.38 | 1.21 | 1.43 |
| regulation of follicle cell microvillus organization | 1 | 3.92 | -1.13 | -1.16 | -1.25 | -1.38 | 1.21 | 1.43 |
| regulation of microvillus length | 1 | 3.92 | -1.13 | -1.16 | -1.25 | -1.38 | 1.21 | 1.43 |
| regulation of follicle cell microvillus length | 1 | 3.92 | -1.13 | -1.16 | -1.25 | -1.38 | 1.21 | 1.43 |
| regulation of cellular component size | 73 | 1.0 | 1.15 | 1.35 | 1.07 | 1.31 | 1.17 | 1.21 |
| regulation of cell projection size | 1 | 3.92 | -1.13 | -1.16 | -1.25 | -1.38 | 1.21 | 1.43 |
| sulfiredoxin activity | 1 | 1.25 | 1.26 | 1.21 | 1.31 | 1.36 | 1.11 | -1.04 |
| mitochondrial translation | 1 | 1.16 | 1.26 | 1.14 | 1.19 | 1.28 | 1.6 | -1.22 |
| ribonucleoside binding | 3 | -1.45 | 1.22 | 1.02 | 1.14 | 1.27 | -1.3 | 1.2 |
| ribonucleotide binding | 807 | 1.08 | 1.1 | 1.19 | 1.08 | 1.22 | 1.17 | 1.09 |
| purine ribonucleotide binding | 807 | 1.08 | 1.1 | 1.19 | 1.08 | 1.22 | 1.17 | 1.09 |
| adenyl ribonucleotide binding | 653 | 1.06 | 1.1 | 1.17 | 1.09 | 1.22 | 1.18 | 1.1 |
| guanyl ribonucleotide binding | 159 | 1.18 | 1.08 | 1.28 | 1.06 | 1.23 | 1.12 | 1.08 |
| O-linoleoyltransferase activity | 1 | -3.36 | 1.4 | 1.32 | 1.25 | 1.35 | -2.25 | 1.21 |
| phosphatidylcholine:cardiolipin O-linoleoyltransferase activity | 1 | -3.36 | 1.4 | 1.32 | 1.25 | 1.35 | -2.25 | 1.21 |
| Golgi cisterna membrane | 3 | 1.17 | 1.23 | -1.14 | 1.08 | -1.09 | 1.82 | 1.37 |
| neuron projection membrane | 1 | -1.08 | -1.3 | -1.04 | 1.04 | 1.56 | 8.15 | 2.89 |
| integral to mitochondrial membrane | 3 | -1.04 | 1.18 | 1.43 | 1.19 | 1.11 | 1.64 | -1.32 |
| regulation of monooxygenase activity | 2 | 1.57 | 1.18 | -1.18 | 1.24 | 1.17 | 1.04 | 1.44 |
| negative regulation of monooxygenase activity | 1 | 1.08 | 1.16 | -1.5 | 1.55 | 1.01 | -2.04 | -1.2 |
| positive regulation of monooxygenase activity | 1 | 2.28 | 1.19 | 1.07 | -1.0 | 1.37 | 2.2 | 2.5 |
| RNA biosynthetic process | 151 | 1.27 | 1.12 | 1.21 | 1.16 | 1.26 | 1.16 | 1.12 |
| Piccolo NuA4 histone acetyltransferase complex | 1 | -1.06 | -1.13 | 1.02 | 1.06 | -1.15 | 1.02 | -1.14 |
| ELL-EAF complex | 8 | 1.25 | 1.11 | 1.53 | 1.07 | 1.44 | 1.46 | 1.18 |
| monocarboxylic acid metabolic process | 78 | 1.06 | -1.1 | -1.24 | -1.08 | -1.06 | -1.06 | -1.03 |
| positive regulation of CREB transcription factor activity | 1 | 2.75 | 1.33 | 1.9 | 1.11 | 2.22 | 3.34 | 2.0 |
| low-density lipoprotein receptor particle metabolic process | 1 | 2.53 | -1.21 | 1.11 | 1.26 | 1.02 | 1.58 | 1.04 |
| carboxy-terminal domain protein kinase complex | 9 | 1.46 | 1.1 | 1.45 | 1.27 | 1.49 | 1.36 | 1.12 |
| tumor necrosis factor receptor superfamily binding | 2 | 1.39 | 1.0 | -1.11 | -1.04 | 2.69 | -1.2 | 2.61 |
| distributive segregation | 6 | 1.23 | -1.03 | -1.06 | 1.1 | 1.19 | 1.15 | 1.25 |
| cell projection cytoplasm | 1 | 1.03 | 1.24 | 1.01 | 1.09 | 1.02 | -1.11 | 1.01 |
| regulation of homeostatic process | 3 | 1.14 | 1.14 | 1.39 | 1.16 | 1.04 | 1.71 | 1.47 |
| positive regulation of Rab GTPase activity | 1 | 1.3 | 1.23 | 1.1 | 1.08 | 1.16 | 2.99 | 1.24 |
| positive regulation of Rac GTPase activity | 2 | -1.11 | 1.35 | 2.32 | 1.27 | 2.42 | 1.41 | 1.46 |
| activation of Ras GTPase activity | 2 | -1.11 | 1.35 | 2.32 | 1.27 | 2.42 | 1.41 | 1.46 |
| activation of Rho GTPase activity | 2 | -1.11 | 1.35 | 2.32 | 1.27 | 2.42 | 1.41 | 1.46 |
| activation of Rac GTPase activity | 2 | -1.11 | 1.35 | 2.32 | 1.27 | 2.42 | 1.41 | 1.46 |
| response to insulin stimulus | 20 | -1.09 | 1.13 | 1.2 | 1.13 | 1.18 | 1.07 | 1.2 |
| cellular response to insulin stimulus | 20 | -1.09 | 1.13 | 1.2 | 1.13 | 1.18 | 1.07 | 1.2 |
| cellular response to hormone stimulus | 52 | 1.07 | 1.12 | 1.17 | 1.08 | 1.23 | 1.19 | 1.21 |
| regulation of stress-activated MAPK cascade | 33 | -1.0 | 1.2 | 1.31 | 1.13 | 1.3 | 1.36 | 1.17 |
| negative regulation of stress-activated MAPK cascade | 12 | 1.09 | 1.13 | 1.44 | 1.11 | 1.23 | 1.18 | 1.08 |
| positive regulation of stress-activated MAPK cascade | 12 | 1.07 | 1.28 | 1.47 | 1.07 | 1.29 | 1.52 | 1.25 |
| regulation of DNA endoreduplication | 5 | -1.5 | 1.1 | 1.17 | 1.11 | 1.42 | -1.56 | 1.43 |
| negative regulation of DNA endoreduplication | 2 | -3.55 | -1.04 | 1.42 | 1.03 | 1.59 | -2.82 | 2.09 |
| positive regulation of DNA endoreduplication | 3 | 1.19 | 1.21 | 1.03 | 1.17 | 1.31 | -1.05 | 1.11 |
| regulation of localization | 163 | 1.09 | 1.07 | 1.21 | 1.06 | 1.19 | 1.26 | 1.15 |
| regulation of protein localization | 48 | 1.04 | 1.11 | 1.27 | 1.11 | 1.24 | 1.15 | 1.16 |
| regulation of polysaccharide metabolic process | 1 | -2.86 | 1.25 | -1.2 | 1.45 | 1.26 | 1.29 | -1.32 |
| regulation of microtubule-based process | 29 | 1.19 | 1.14 | 1.41 | 1.06 | 1.41 | 1.52 | 1.27 |
| regulation of spindle elongation | 1 | -1.34 | -1.01 | 1.11 | 1.33 | 1.45 | -1.15 | -1.3 |
| regulation of mitotic spindle elongation | 1 | -1.34 | -1.01 | 1.11 | 1.33 | 1.45 | -1.15 | -1.3 |
| circadian regulation of gene expression | 3 | 1.48 | 1.24 | 1.41 | 1.04 | 1.37 | 1.9 | 1.48 |
| activin receptor signaling pathway | 3 | -2.1 | 2.02 | -1.27 | -1.55 | -1.26 | -1.31 | 1.66 |
| regulation of activin receptor signaling pathway | 1 | 1.88 | -1.02 | -1.08 | -1.19 | -1.0 | 2.96 | -1.02 |
| negative regulation of activin receptor signaling pathway | 1 | 1.88 | -1.02 | -1.08 | -1.19 | -1.0 | 2.96 | -1.02 |
| positive regulation of activin receptor signaling pathway | 1 | 1.88 | -1.02 | -1.08 | -1.19 | -1.0 | 2.96 | -1.02 |
| SREBP-mediated signaling pathway | 6 | 1.15 | 1.2 | 1.22 | 1.13 | 1.33 | -1.04 | 1.13 |
| sterol binding | 17 | -2.71 | -1.57 | -1.93 | 1.12 | -1.21 | -2.6 | -2.64 |
| secretion by cell | 139 | -1.02 | 1.06 | 1.22 | 1.11 | 1.23 | 1.11 | 1.08 |
| protein complex scaffold | 2 | 1.19 | 1.14 | 1.28 | -1.02 | 1.24 | 1.13 | 1.22 |
| regulation of cytokinetic process | 1 | -1.32 | 1.18 | 1.39 | 1.03 | 1.12 | 1.4 | 1.35 |
| regulation of actin cytoskeleton organization | 30 | -1.3 | 1.08 | 1.43 | 1.03 | 1.44 | -1.08 | 1.19 |
| inositol phosphate biosynthetic process | 3 | -1.14 | 1.2 | -1.03 | -1.06 | 1.01 | -1.08 | 1.11 |
| positive regulation of transcription elongation from RNA polymerase II promoter | 10 | -1.01 | 1.29 | 1.32 | -1.05 | 1.22 | 1.32 | 1.65 |
| regulation of actin filament-based process | 30 | -1.3 | 1.08 | 1.43 | 1.03 | 1.44 | -1.08 | 1.19 |
| mitochondrial respiratory chain complex I assembly | 1 | 1.08 | 1.05 | 1.31 | 1.03 | 1.07 | 1.99 | -1.52 |
| myosin filament | 3 | 2.91 | -1.49 | 1.05 | -1.32 | 1.04 | 2.39 | -1.29 |
| macromolecular complex disassembly | 16 | 1.02 | 1.16 | 1.29 | 1.14 | 1.31 | 1.19 | 1.04 |
| ribonucleoprotein complex disassembly | 1 | 1.69 | 1.29 | 1.56 | 1.32 | 1.6 | 1.63 | 1.44 |
| cellular component morphogenesis | 513 | 1.16 | 1.08 | 1.23 | 1.03 | 1.24 | 1.24 | 1.21 |
| cell part morphogenesis | 359 | 1.11 | 1.08 | 1.23 | 1.04 | 1.24 | 1.23 | 1.23 |
| macromolecular complex | 1903 | 1.15 | 1.08 | 1.19 | 1.09 | 1.19 | 1.19 | 1.03 |
| protein-carbohydrate complex | 1 | 2.19 | 1.14 | 2.0 | 1.19 | 2.09 | 1.71 | 1.7 |
| protein-DNA complex | 35 | -1.03 | 1.07 | 1.21 | 1.2 | 1.27 | -1.15 | -1.1 |
| tetrapyrrole metabolic process | 13 | 1.13 | 1.17 | 1.08 | 1.1 | -1.02 | 1.13 | -1.08 |
| tetrapyrrole biosynthetic process | 12 | 1.08 | 1.18 | 1.02 | 1.11 | -1.05 | 1.06 | -1.1 |
| tetrapyrrole catabolic process | 1 | 2.0 | 1.05 | 2.09 | 1.06 | 1.46 | 2.34 | 1.12 |
| bitter taste receptor activity | 2 | -1.31 | -1.2 | -1.22 | -1.24 | 3.17 | 1.22 | -1.17 |
| sweet taste receptor activity | 2 | -1.42 | -1.09 | -1.0 | 1.18 | -1.03 | -1.26 | -1.15 |
| regulation of organelle organization | 124 | 1.1 | 1.1 | 1.3 | 1.07 | 1.34 | 1.17 | 1.17 |
| regulation of chromosome organization | 15 | 1.16 | 1.17 | 1.39 | 1.18 | 1.42 | 1.32 | 1.27 |
| multicellular organismal reproductive behavior | 73 | -1.01 | -1.11 | -1.02 | -1.04 | -1.08 | -1.0 | 1.04 |
| cellular pigmentation | 11 | 1.07 | 1.19 | 1.37 | 1.13 | 1.22 | 1.15 | 1.17 |
| ocellus pigmentation | 18 | 1.35 | 1.13 | 1.12 | 1.06 | 1.06 | 1.08 | 1.16 |
| mitochondrial respiratory chain complex assembly | 2 | 1.06 | 1.05 | 1.15 | -1.01 | -1.01 | 1.5 | -1.3 |
| negative regulation of RNA splicing | 5 | 1.21 | 1.27 | 1.23 | 1.22 | 1.16 | 1.36 | 1.08 |
| regulation of purine nucleotide catabolic process | 43 | 1.08 | 1.16 | 1.36 | 1.06 | 1.3 | 1.31 | 1.21 |
| regulation of GTP catabolic process | 42 | 1.08 | 1.16 | 1.35 | 1.05 | 1.29 | 1.31 | 1.21 |
| acetylcholine receptor binding | 1 | -1.02 | 1.16 | 1.08 | 1.02 | 1.02 | 1.28 | 1.01 |
| regulation of intracellular protein transport | 35 | 1.17 | 1.1 | 1.23 | 1.1 | 1.24 | 1.22 | 1.23 |
| regulation of protein import into nucleus, translocation | 2 | -1.04 | 1.19 | 1.3 | 1.1 | -1.02 | 1.0 | 1.13 |
| positive regulation of protein import into nucleus, translocation | 2 | -1.04 | 1.19 | 1.3 | 1.1 | -1.02 | 1.0 | 1.13 |
| conversion of ds siRNA to ss siRNA involved in RNA interference | 1 | 1.02 | 1.47 | 1.88 | 1.06 | 1.63 | 2.09 | 1.83 |
| histone H3-K9 demethylation | 2 | 1.08 | 1.24 | 1.45 | 1.25 | 1.55 | 1.06 | 1.05 |
| protein-DNA loading ATPase activity | 3 | 1.23 | 1.05 | 1.23 | 1.24 | 1.24 | 1.1 | -1.01 |
| proton-transporting V-type ATPase complex | 28 | -1.62 | -1.0 | 1.32 | -1.18 | -1.18 | 1.26 | -1.13 |
| proton-transporting two-sector ATPase complex, proton-transporting domain | 24 | -1.5 | 1.0 | 1.3 | -1.13 | -1.09 | 1.2 | -1.21 |
| proton-transporting two-sector ATPase complex, catalytic domain | 24 | 1.11 | -1.0 | 1.24 | -1.09 | -1.02 | 1.63 | -1.09 |
| proton-transporting V-type ATPase, V0 domain | 15 | -2.14 | -1.02 | 1.29 | -1.23 | -1.29 | 1.0 | -1.25 |
| proton-transporting V-type ATPase, V1 domain | 14 | -1.13 | 1.02 | 1.36 | -1.11 | -1.05 | 1.59 | 1.0 |
| response to hydroperoxide | 1 | 2.0 | -1.09 | -1.07 | 1.14 | 1.47 | 1.02 | -1.28 |
| DNA helicase complex | 9 | -1.14 | 1.07 | 1.16 | 1.17 | 1.25 | -1.1 | -1.02 |
| cell cycle cytokinesis | 34 | 1.28 | 1.02 | 1.18 | -1.01 | 1.3 | 1.18 | 1.14 |
| cytokinesis after meiosis | 19 | 1.55 | 1.01 | 1.2 | -1.08 | 1.35 | 1.45 | 1.26 |
| iron assimilation | 1 | -2.45 | 1.57 | -1.18 | 1.13 | -1.31 | -2.1 | 1.39 |
| dsRNA transport | 22 | -1.32 | 1.18 | 1.23 | 1.09 | 1.33 | 1.1 | 1.01 |
| regulation of cellular amine metabolic process | 3 | 1.05 | 1.06 | 1.07 | 1.15 | 1.1 | -1.22 | 1.43 |
| regulation of S phase | 16 | 1.38 | 1.18 | 1.34 | 1.13 | 1.4 | 1.63 | 1.43 |
| axon part | 31 | 1.25 | 1.05 | 1.11 | -1.11 | 1.16 | 1.72 | 1.28 |
| monocarboxylic acid binding | 11 | -1.61 | -1.59 | -2.07 | -1.15 | 1.3 | -1.68 | -1.56 |
| cell cycle comprising mitosis without cytokinesis | 14 | 1.58 | -1.04 | 1.19 | -1.07 | 1.25 | 1.21 | 1.5 |
| meiotic cell cycle checkpoint | 1 | 1.5 | 1.02 | 1.68 | -1.05 | 1.78 | 1.86 | 1.49 |
| protein localization to organelle | 102 | 1.05 | 1.19 | 1.2 | 1.14 | 1.18 | 1.07 | 1.13 |
| pronuclear migration | 4 | -1.16 | 1.07 | 1.08 | 1.03 | 1.23 | -1.25 | -1.05 |
| carbohydrate homeostasis | 5 | 1.14 | 1.16 | -1.21 | -1.59 | -1.34 | 1.07 | 1.69 |
| fatty acid beta-oxidation using acyl-CoA oxidase | 1 | -1.23 | 1.06 | -2.67 | 1.58 | 1.11 | -5.4 | -1.98 |
| MAP kinase phosphatase activity | 3 | 1.18 | 1.17 | 1.3 | 1.08 | 1.35 | 1.14 | 1.33 |
| multicellular organismal response to stress | 2 | 1.88 | -1.18 | 1.22 | 1.16 | 1.43 | 1.98 | 1.19 |
| Slx1-Slx4 complex | 1 | 1.24 | 1.01 | -1.1 | -1.01 | -1.04 | 1.25 | -1.13 |
| protein deacetylase activity | 8 | 1.29 | 1.14 | 1.07 | 1.21 | 1.24 | 1.41 | 1.17 |
| unsaturated fatty acid metabolic process | 1 | -1.74 | 1.11 | -2.01 | 1.51 | 1.27 | -1.87 | -1.55 |
| dorsal/ventral axon guidance | 2 | 1.25 | 1.19 | 1.54 | 1.04 | 1.22 | 1.73 | 1.43 |
| protein glycosylation in endoplasmic reticulum | 1 | 2.78 | -1.05 | 1.02 | -1.07 | 1.12 | 2.02 | 1.06 |
| protein glycosylation in Golgi | 1 | 1.63 | 1.17 | -1.53 | -1.05 | -1.49 | 4.17 | 1.53 |
| rhabdomere membrane | 1 | 1.12 | 1.01 | 1.02 | -1.01 | 1.1 | -1.08 | -1.09 |
| Elongator holoenzyme complex | 1 | -1.53 | 1.18 | -1.12 | 1.34 | -1.02 | -1.63 | -1.29 |
| activating transcription factor binding | 9 | 1.49 | -1.18 | -1.05 | -1.08 | 1.1 | 1.31 | 1.18 |
| mitochondrial proton-transporting ATP synthase complex assembly | 1 | 1.02 | 1.07 | 1.3 | 1.18 | 1.05 | 1.56 | -1.57 |
| membrane protein proteolysis | 4 | -1.09 | 1.28 | 1.16 | 1.23 | 1.32 | -1.25 | 1.31 |
| cell adhesion mediated by integrin | 2 | -1.31 | -1.12 | -1.25 | -1.23 | -1.34 | -1.41 | -1.27 |
| regulation of cell adhesion mediated by integrin | 2 | -1.34 | 1.03 | 1.08 | -1.01 | 1.21 | -1.37 | 1.03 |
| regulation of growth of symbiont in host | 7 | 2.04 | -1.02 | 1.37 | 1.07 | 1.52 | 1.47 | 1.08 |
| negative regulation of growth of symbiont in host | 7 | 2.04 | -1.02 | 1.37 | 1.07 | 1.52 | 1.47 | 1.08 |
| negative regulation of kinase activity | 12 | 1.53 | 1.14 | 1.46 | 1.09 | 1.41 | 1.69 | 1.32 |
| positive regulation of kinase activity | 25 | 1.18 | 1.14 | 1.29 | 1.1 | 1.23 | 1.49 | 1.19 |
| nucleotide-excision repair, DNA incision | 1 | 1.98 | 1.16 | 1.33 | 1.18 | 1.39 | 1.07 | 1.02 |
| cellular polysaccharide biosynthetic process | 11 | 1.69 | 1.06 | -1.03 | 1.08 | 1.23 | 1.68 | 1.33 |
| establishment of ribosome localization | 1 | 2.15 | -1.01 | 1.44 | 1.25 | 1.44 | 1.28 | 1.21 |
| steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 3 | 2.12 | -1.18 | -1.46 | -1.03 | -1.03 | 1.26 | -1.06 |
| steroid dehydrogenase activity, acting on the CH-CH group of donors | 1 | -1.31 | 1.28 | -1.32 | 1.3 | 1.08 | -2.32 | -1.18 |
| SUMO-targeted ubiquitin ligase complex | 1 | 1.3 | 1.31 | 1.72 | 1.5 | 1.61 | 1.9 | 1.2 |
| O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity | 1 | 1.31 | 1.46 | -1.17 | -1.24 | 1.74 | -1.95 | 3.54 |
| N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity | 2 | -1.04 | 1.82 | 1.08 | 1.06 | -1.12 | -1.39 | 1.31 |
| nucleoside bisphosphate metabolic process | 6 | 1.42 | 1.19 | 1.1 | 1.23 | 1.41 | -1.05 | -1.06 |
| ribonuclease T2 activity | 1 | 2.66 | -1.03 | 1.81 | 1.01 | 1.48 | 1.86 | 1.52 |
| mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase activity | 1 | -1.77 | -1.11 | 1.92 | 1.18 | 1.33 | -1.27 | -1.19 |
| cis-stilbene-oxide hydrolase activity | 3 | -2.11 | 1.3 | -1.14 | -1.06 | -1.44 | -4.35 | 1.38 |
| cytoplasmic mRNA processing body assembly | 5 | 1.3 | 1.16 | 1.47 | 1.28 | 1.54 | 1.31 | 1.13 |
| 5-(carboxyamino)imidazole ribonucleotide mutase activity | 1 | 1.05 | -1.55 | -1.44 | -1.5 | -1.3 | 3.7 | 1.09 |
| endosomal vesicle fusion | 4 | 1.19 | 1.07 | 2.26 | 1.26 | 1.71 | 1.76 | 1.23 |
| response to anoxia | 5 | 1.07 | 1.07 | 1.2 | 1.15 | 1.24 | 1.28 | 1.15 |
| DNA polymerase activity | 19 | 1.07 | 1.12 | 1.2 | 1.07 | 1.19 | 1.1 | -1.04 |
| RNA polymerase activity | 28 | 1.31 | 1.12 | 1.19 | 1.24 | 1.24 | 1.06 | 1.0 |
| cellular response to amino acid starvation | 4 | -1.04 | 1.16 | 1.31 | 1.01 | 1.16 | 1.47 | 1.31 |
| peptide N-acetyltransferase activity | 4 | 1.05 | 1.16 | 1.37 | 1.09 | 1.14 | 1.03 | 1.01 |
| ion transmembrane transport | 73 | -1.07 | 1.02 | 1.16 | -1.11 | -1.07 | 1.38 | -1.11 |
| GPI anchor binding | 1 | -1.1 | 1.17 | -1.13 | -1.04 | -1.08 | -1.08 | 1.11 |
| regulation of transcription elongation from RNA polymerase II promoter | 14 | 1.05 | 1.24 | 1.29 | 1.01 | 1.21 | 1.26 | 1.46 |
| negative regulation of transcription elongation from RNA polymerase II promoter | 4 | 1.21 | 1.13 | 1.22 | 1.16 | 1.17 | 1.14 | 1.08 |
| response to monosaccharide stimulus | 2 | -1.1 | 1.06 | 1.16 | 1.15 | 1.11 | -1.04 | -1.08 |
| response to disaccharide stimulus | 5 | 2.19 | -1.05 | 1.25 | 1.03 | 1.35 | 1.69 | 1.35 |
| response to maltose stimulus | 1 | -1.49 | -1.13 | 1.17 | 1.11 | 1.02 | -1.25 | -1.24 |
| detection of monosaccharide stimulus | 1 | -1.49 | -1.13 | 1.17 | 1.11 | 1.02 | -1.25 | -1.24 |
| detection of disaccharide stimulus | 1 | -1.49 | -1.13 | 1.17 | 1.11 | 1.02 | -1.25 | -1.24 |
| detection of maltose stimulus | 1 | -1.49 | -1.13 | 1.17 | 1.11 | 1.02 | -1.25 | -1.24 |
| primary alcohol metabolic process | 2 | 1.95 | -1.28 | -2.91 | -1.89 | 1.06 | 1.72 | 1.15 |
| diol metabolic process | 9 | -1.02 | -1.18 | -1.26 | -1.16 | -1.28 | -1.1 | 1.52 |
| diol biosynthetic process | 3 | -1.04 | -2.12 | -1.47 | -1.4 | -1.53 | 1.41 | 2.4 |
| diol catabolic process | 1 | -8.98 | 1.77 | -2.05 | -1.02 | -2.08 | -14.22 | 1.22 |
| Arp2/3 complex-mediated actin nucleation | 2 | -1.4 | 1.24 | 1.37 | 1.14 | 1.19 | -1.91 | 1.02 |
| cell junction assembly | 37 | 1.87 | -1.1 | 1.3 | -1.15 | 1.23 | 2.13 | 1.42 |
| cell junction organization | 46 | 1.6 | -1.06 | 1.34 | -1.1 | 1.25 | 1.87 | 1.32 |
| cell junction maintenance | 9 | -1.01 | 1.16 | 1.73 | 1.07 | 1.49 | 1.28 | 1.13 |
| adherens junction organization | 12 | 1.06 | 1.11 | 1.42 | 1.06 | 1.32 | 1.17 | 1.09 |
| adherens junction assembly | 2 | -1.2 | -1.04 | -1.19 | 1.02 | -1.08 | -1.21 | -1.15 |
| adherens junction maintenance | 7 | -1.01 | 1.13 | 1.67 | 1.07 | 1.42 | 1.13 | -1.02 |
| macromolecular complex remodeling | 5 | 1.21 | 1.11 | 1.42 | 1.33 | 1.33 | 1.15 | 1.09 |
| lipid particle organization | 6 | 1.5 | -1.3 | -1.62 | 1.09 | 1.28 | -1.38 | 1.28 |
| nuclear periphery | 14 | 1.25 | 1.15 | 1.12 | 1.25 | 1.18 | 1.1 | 1.05 |
| nuclear-transcribed mRNA catabolic process, exonucleolytic, 5'-3' | 1 | -1.45 | 1.76 | 1.14 | 1.42 | 1.29 | -1.3 | 1.32 |
| lipid oxidation | 14 | -1.0 | 1.05 | -1.49 | 1.08 | 1.25 | -1.91 | -1.22 |
| ubiquitin-ubiquitin ligase activity | 2 | 1.19 | 1.09 | 1.35 | 1.16 | 1.35 | 1.16 | 1.36 |
| microtubule anchoring | 6 | -1.07 | 1.06 | 1.1 | 1.13 | 1.24 | 1.02 | -1.0 |
| microtubule anchoring at centrosome | 1 | -1.51 | -1.02 | 1.38 | 1.04 | 1.42 | 1.11 | -1.1 |
| ncRNA processing | 64 | 1.28 | 1.14 | 1.2 | 1.24 | 1.22 | 1.11 | 1.01 |
| chondroitin sulfotransferase activity | 2 | -1.02 | 1.22 | 1.17 | 1.01 | 1.0 | 1.01 | -1.01 |
| heparan sulfate sulfotransferase activity | 6 | 1.58 | 1.12 | 1.54 | 1.02 | 1.18 | 1.11 | 1.46 |
| protein localization to kinetochore | 1 | 2.47 | 1.54 | 1.18 | 1.04 | 1.48 | 1.78 | 1.43 |
| protein localization to chromosome | 1 | 2.47 | 1.54 | 1.18 | 1.04 | 1.48 | 1.78 | 1.43 |
| protein localization to nucleus | 38 | 1.16 | 1.19 | 1.34 | 1.21 | 1.35 | 1.22 | 1.15 |
| centromere complex assembly | 3 | 1.0 | 1.03 | -1.07 | 1.16 | 1.05 | -1.12 | -1.1 |
| box C/D snoRNA binding | 1 | 1.11 | -1.14 | 1.0 | 1.3 | 1.17 | -1.17 | -1.17 |
| RNA cap binding complex | 4 | 1.34 | 1.15 | 1.34 | 1.16 | 1.42 | 1.43 | 1.2 |
| mitochondrial respiratory chain complex III assembly | 1 | 1.04 | 1.04 | 1.01 | -1.04 | -1.1 | 1.13 | -1.12 |
| piRNA binding | 2 | 1.58 | 1.07 | 1.4 | 1.11 | 1.03 | 1.55 | 1.05 |
| phosphatidylinositol phosphate 4-phosphatase activity | 1 | -1.07 | 1.22 | 1.09 | 1.37 | -1.04 | -1.17 | -1.02 |
| cellular response to oxidative stress | 17 | 1.07 | -1.16 | -1.22 | -1.13 | -1.0 | 1.33 | -1.03 |
| cellular response to heat | 20 | -1.11 | -1.25 | -1.29 | -1.41 | -1.07 | 1.17 | 1.13 |
| cellular protein localization | 214 | 1.06 | 1.16 | 1.29 | 1.13 | 1.23 | 1.14 | 1.17 |
| cellular response to reactive oxygen species | 11 | -1.03 | 1.06 | 1.07 | 1.16 | 1.2 | -1.19 | -1.14 |
| cellular response to unfolded protein | 1 | -1.05 | 1.27 | -1.17 | -1.24 | -1.49 | -1.01 | 1.19 |
| cellular macromolecular complex subunit organization | 173 | 1.07 | 1.1 | 1.28 | 1.14 | 1.27 | 1.11 | 1.03 |
| cellular macromolecular complex assembly | 143 | 1.05 | 1.09 | 1.28 | 1.14 | 1.27 | 1.07 | 1.01 |
| cellular macromolecular complex disassembly | 16 | 1.02 | 1.16 | 1.29 | 1.14 | 1.31 | 1.19 | 1.04 |
| cellular protein complex localization | 2 | 2.1 | 1.14 | 1.17 | 1.2 | 1.38 | 1.71 | 1.06 |
| cellular carbohydrate biosynthetic process | 22 | 1.52 | -1.16 | -1.16 | -1.14 | 1.15 | 1.68 | 1.12 |
| cellular nitrogen compound metabolic process | 1149 | 1.16 | 1.09 | 1.14 | 1.11 | 1.19 | 1.16 | 1.05 |
| establishment of mitochondrion localization, microtubule-mediated | 5 | -1.72 | 1.29 | 1.54 | 1.18 | 1.39 | 1.16 | 1.27 |
| cellular response to UV | 11 | -1.5 | -1.74 | -2.0 | -1.64 | -1.56 | -1.07 | -1.13 |
| cellular macromolecule biosynthetic process | 627 | 1.18 | 1.09 | 1.17 | 1.11 | 1.21 | 1.19 | 1.03 |
| histone demethylase activity (H3-trimethyl-K4 specific) | 2 | 1.08 | 1.42 | 1.76 | 1.01 | 1.58 | 2.53 | 1.88 |
| nucleobase-containing compound biosynthetic process | 98 | 1.15 | 1.06 | 1.08 | 1.01 | 1.06 | 1.29 | -1.01 |
| nucleobase-containing compound catabolic process | 55 | -1.05 | 1.01 | -1.01 | 1.05 | 1.04 | -1.13 | 1.03 |
| ncRNA metabolic process | 107 | 1.13 | 1.19 | 1.23 | 1.2 | 1.21 | 1.08 | 1.04 |
| ncRNA catabolic process | 1 | 1.49 | 1.15 | 1.39 | 1.19 | 1.14 | 2.46 | 1.32 |
| tripeptidase activity | 1 | -2.16 | -1.91 | -1.31 | 1.97 | 1.89 | -1.93 | -2.32 |
| ion channel complex | 41 | -1.0 | 1.04 | 1.08 | -1.03 | -1.06 | -1.06 | -1.04 |
| cation channel complex | 25 | 1.07 | 1.02 | 1.08 | 1.02 | 1.04 | -1.01 | -1.04 |
| calcium channel complex | 4 | 1.36 | -1.08 | -1.02 | 1.02 | -1.08 | 1.17 | -1.12 |
| potassium channel complex | 18 | 1.03 | 1.03 | 1.1 | 1.02 | 1.05 | -1.06 | -1.0 |
| sodium channel complex | 1 | 1.49 | 1.1 | 1.18 | 1.37 | 1.22 | 1.19 | 1.04 |
| chloride channel complex | 6 | -1.41 | 1.18 | 1.24 | -1.25 | -1.53 | -1.47 | -1.09 |
| methyltransferase complex | 16 | 1.51 | 1.16 | 1.4 | 1.13 | 1.36 | 1.38 | 1.21 |
| histone H3-K4 demethylation | 3 | 1.04 | 1.4 | 1.66 | 1.06 | 1.46 | 1.94 | 1.65 |
| histone H3-K4 demethylation, trimethyl-H3-K4-specific | 1 | 1.33 | 1.27 | 1.61 | -1.08 | 1.55 | 2.66 | 1.67 |
| DNA replication-independent nucleosome organization | 1 | -1.49 | 1.47 | -1.04 | 1.39 | 1.42 | -1.65 | -1.06 |
| cellular hormone metabolic process | 22 | 1.12 | -1.16 | -1.13 | -1.16 | -1.01 | 1.05 | 1.21 |
| regulation of transmembrane transport | 35 | 1.19 | 1.08 | 1.22 | 1.07 | 1.22 | 1.3 | 1.2 |
| negative regulation of transmembrane transport | 6 | 1.84 | 1.13 | 1.25 | 1.03 | 1.14 | 1.94 | 1.57 |
| positive regulation of transmembrane transport | 18 | 1.13 | 1.06 | 1.22 | 1.1 | 1.37 | 1.17 | 1.13 |
| regulation of ion transmembrane transport | 8 | 1.33 | -1.01 | 1.17 | -1.03 | 1.1 | 1.52 | 1.12 |
| negative regulation of ion transmembrane transport | 1 | -1.32 | 1.16 | 1.21 | -1.14 | -1.21 | 1.47 | 1.08 |
| basement membrane disassembly | 3 | -2.47 | 1.3 | 1.22 | 1.11 | 1.41 | -1.09 | 1.18 |
| histone H4-K20 methylation | 3 | 1.22 | 1.28 | 1.67 | 1.2 | 1.75 | 1.82 | 1.4 |
| histone H4-K20 monomethylation | 1 | 1.22 | 1.34 | 1.86 | 1.14 | 2.08 | 2.98 | 1.75 |
| histone H4-K20 dimethylation | 1 | 1.41 | 1.59 | 1.77 | 1.23 | 1.83 | 1.65 | 1.69 |
| histone H4-K20 trimethylation | 2 | 1.21 | 1.25 | 1.58 | 1.24 | 1.6 | 1.43 | 1.25 |
| histone lysine methylation | 19 | 1.48 | 1.18 | 1.43 | 1.13 | 1.43 | 1.45 | 1.28 |
| histone arginine methylation | 2 | 1.3 | 1.22 | 1.14 | 1.25 | 1.27 | 1.38 | 1.31 |
| response to endoplasmic reticulum stress | 3 | -1.16 | 1.37 | 1.28 | -1.02 | 1.1 | -1.11 | 1.57 |
| NAD-dependent protein deacetylase activity | 2 | 1.85 | 1.16 | 1.29 | 1.09 | 1.23 | 1.67 | 1.46 |
| mitochondrial protein processing | 2 | 1.16 | 1.04 | 1.32 | 1.16 | 1.03 | 1.09 | -1.27 |
| iron chaperone activity | 1 | 1.38 | 1.05 | 1.33 | 1.44 | 1.46 | 1.81 | -1.29 |
| dorsal trunk growth, open tracheal system | 4 | 1.59 | -1.02 | 1.16 | -1.15 | 1.19 | 1.9 | 1.23 |
| liquid clearance, open tracheal system | 6 | 1.48 | 1.17 | 1.29 | 1.11 | 1.28 | 1.82 | 1.45 |
| subapical complex | 13 | 1.4 | 1.1 | 1.14 | 1.01 | 1.29 | 2.02 | 1.24 |
| phosphatidylinositol 3-kinase activity | 3 | -1.46 | 1.15 | 1.53 | 1.18 | 1.77 | -1.09 | -1.14 |
| melanization defense response | 12 | 2.45 | -1.25 | -1.18 | -1.23 | 1.2 | 1.6 | 1.51 |
| regulation of melanization defense response | 3 | 10.08 | -1.98 | -1.97 | -2.05 | -1.49 | 4.83 | 1.41 |
| positive regulation of melanization defense response | 1 | 13.93 | -2.44 | -2.81 | -2.18 | -1.62 | 11.32 | -1.11 |
| negative regulation of melanization defense response | 1 | 23.01 | -3.14 | -2.9 | -3.17 | -2.48 | 1.73 | 2.58 |
| encapsulation of foreign target | 6 | 2.07 | -1.07 | 1.2 | -1.17 | 1.1 | 1.42 | 1.52 |
| melanotic encapsulation of foreign target | 2 | 5.77 | -1.55 | -1.53 | -1.72 | -1.45 | 1.51 | 1.76 |
| polytene chromosome, telomeric region | 5 | 1.25 | 1.13 | 1.41 | 1.18 | 1.43 | 1.46 | 1.14 |
| myosuppressin receptor activity | 1 | 13.14 | -1.35 | -1.29 | -1.22 | -1.17 | -1.49 | -1.27 |
| phosphatidylinositol 3-kinase regulator activity | 1 | -2.08 | 1.8 | 1.08 | 1.22 | 1.65 | -1.22 | 1.26 |
| elongation of arista core | 2 | -1.25 | 1.12 | -1.06 | -1.0 | -1.17 | -1.12 | -1.24 |
| cuticle pattern formation | 23 | 1.0 | 1.04 | 1.21 | -1.06 | 1.14 | 1.06 | 1.51 |
| adult chitin-based cuticle pattern formation | 1 | -1.38 | -1.23 | -1.19 | -1.18 | -1.36 | -1.47 | -1.32 |
| somatic stem cell maintenance | 13 | 1.07 | 1.2 | 1.28 | 1.19 | 1.34 | 1.15 | 1.3 |
| regulation of Rac protein signal transduction | 2 | 1.51 | 1.07 | 1.28 | -1.16 | 1.49 | 6.25 | 1.07 |
| negative regulation of Rac protein signal transduction | 1 | 2.11 | 1.3 | 1.76 | 1.16 | 2.29 | 4.26 | 1.69 |
| regulation of Rho protein signal transduction | 26 | 1.09 | 1.14 | 1.48 | 1.09 | 1.46 | 1.56 | 1.26 |
| positive regulation of Rho protein signal transduction | 1 | 2.11 | 1.3 | 1.76 | 1.16 | 2.29 | 4.26 | 1.69 |
| leading edge cell differentiation | 5 | 1.03 | 1.07 | 2.19 | -1.04 | 1.49 | -1.17 | 1.24 |
| leading edge cell fate commitment | 3 | 1.0 | 1.08 | 1.89 | 1.0 | 1.21 | -1.44 | -1.04 |
| leading edge cell fate determination | 3 | 1.0 | 1.08 | 1.89 | 1.0 | 1.21 | -1.44 | -1.04 |
| dorsal closure, leading edge cell fate commitment | 3 | 1.0 | 1.08 | 1.89 | 1.0 | 1.21 | -1.44 | -1.04 |
| phosphatidylinositol 3-kinase complex, class III | 1 | -1.02 | 1.18 | 1.13 | 1.36 | 1.14 | 1.29 | -1.24 |
| female pronucleus assembly | 2 | 1.19 | 1.0 | 1.34 | 1.1 | 1.22 | 1.11 | -1.19 |
| male pronucleus assembly | 4 | -1.1 | 1.36 | 1.12 | 1.11 | -1.05 | 1.25 | 1.15 |
| sperm chromatin decondensation | 3 | -1.17 | 1.37 | 1.22 | 1.18 | 1.07 | -1.09 | 1.13 |
| fertilization, exchange of chromosomal proteins | 3 | -1.17 | 1.37 | 1.22 | 1.18 | 1.07 | -1.09 | 1.13 |
| sperm aster formation | 4 | -1.22 | 1.05 | -1.0 | 1.07 | 1.17 | -1.34 | -1.21 |
| sperm plasma membrane disassembly | 1 | -1.22 | 1.14 | 1.06 | 1.01 | -1.52 | -1.37 | 1.04 |
| splicing factor protein import into nucleus | 1 | -1.2 | 1.18 | -1.09 | 1.33 | -1.05 | -1.15 | -1.23 |
| juvenile hormone acid methyltransferase activity | 1 | 2.57 | -9.13 | -9.92 | -9.08 | -2.93 | -2.21 | 6.69 |
| embryonic heart tube development | 14 | -1.03 | 1.04 | 1.18 | 1.02 | 1.18 | -1.05 | 1.47 |
| cardiac cell differentiation | 20 | 1.36 | -1.01 | 1.21 | -1.08 | 1.08 | 1.53 | 1.21 |
| dorsal vessel aortic cell fate commitment | 1 | -1.23 | 1.26 | -1.02 | -1.1 | -1.2 | 1.25 | -1.03 |
| dorsal vessel heart proper cell fate commitment | 2 | 1.33 | 1.12 | 1.13 | 1.09 | 1.14 | 1.5 | 1.21 |
| embryonic heart tube anterior/posterior pattern specification | 3 | 1.13 | 1.16 | 1.08 | 1.02 | 1.03 | 1.41 | 1.13 |
| nonmotile primary cilium assembly | 11 | -1.06 | 1.06 | -1.03 | 1.1 | 1.07 | 1.03 | -1.06 |
| RCAF complex | 1 | -1.52 | 1.22 | 1.31 | 1.37 | 1.19 | -1.61 | -1.48 |
| brahma complex | 9 | 1.16 | 1.17 | 1.42 | 1.17 | 1.47 | 1.4 | 1.29 |
| omega speckle | 4 | -1.04 | 1.25 | 1.21 | 1.21 | 1.23 | 1.2 | -1.07 |
| nuclear speck organization | 1 | 1.08 | 1.17 | 2.37 | 1.04 | 2.17 | 1.91 | 1.84 |
| methylated histone residue binding | 6 | 1.3 | 1.14 | 1.32 | 1.26 | 1.24 | 1.32 | 1.04 |
| regulation of histone acetylation | 5 | 1.41 | 1.17 | 1.61 | 1.2 | 1.61 | 1.62 | 1.46 |
| positive regulation of histone acetylation | 2 | 1.41 | 1.2 | 1.81 | 1.29 | 1.46 | 1.33 | 1.45 |
| negative regulation of histone acetylation | 1 | 1.22 | 1.34 | 1.86 | 1.14 | 2.08 | 2.98 | 1.75 |
| micro-ribonucleoprotein complex | 1 | -1.57 | 1.18 | 1.13 | 1.01 | 1.08 | -1.1 | 1.28 |
| larval midgut histolysis | 8 | -1.09 | 1.19 | 1.22 | 1.19 | 1.25 | 1.0 | 1.3 |
| salivary gland histolysis | 73 | -1.0 | 1.0 | 1.1 | 1.06 | 1.24 | 1.15 | 1.1 |
| salivary gland cell autophagic cell death | 70 | 1.04 | -1.0 | 1.1 | 1.06 | 1.23 | 1.21 | 1.11 |
| ecdysone-mediated induction of salivary gland cell autophagic cell death | 11 | 1.07 | 1.06 | 1.25 | 1.02 | 1.27 | 1.36 | 1.36 |
| pupariation | 7 | 1.31 | 1.04 | 1.24 | -1.05 | 1.2 | 2.66 | 1.16 |
| response to ecdysone | 27 | 1.12 | 1.07 | 1.54 | 1.02 | 1.35 | 1.59 | 1.26 |
| ecdysone receptor-mediated signaling pathway | 8 | 1.35 | 1.22 | 1.56 | 1.09 | 1.48 | 1.56 | 1.37 |
| induction of programmed cell death by ecdysone | 12 | 1.05 | 1.07 | 1.28 | 1.03 | 1.29 | 1.32 | 1.33 |
| polytene chromosome puffing | 3 | 1.97 | -1.0 | -1.28 | -1.11 | 1.05 | -1.03 | 2.22 |
| heat shock-mediated polytene chromosome puffing | 3 | 1.97 | -1.0 | -1.28 | -1.11 | 1.05 | -1.03 | 2.22 |
| induction of programmed cell death by hormones | 13 | 1.07 | 1.08 | 1.33 | 1.05 | 1.34 | 1.34 | 1.34 |
| axoneme assembly | 9 | -1.07 | 1.1 | 1.21 | 1.07 | 1.11 | -1.04 | -1.0 |
| cilium axoneme assembly | 2 | -1.34 | 1.15 | 1.69 | 1.06 | -1.04 | -1.19 | -1.09 |
| flagellar axoneme assembly | 7 | -1.01 | 1.09 | 1.1 | 1.07 | 1.15 | -1.0 | 1.02 |
| flagellar axoneme | 1 | 1.03 | 1.24 | 1.01 | 1.09 | 1.02 | -1.11 | 1.01 |
| siRNA loading onto RISC involved in RNA interference | 4 | 1.18 | 1.15 | 1.71 | 1.01 | 1.53 | 1.21 | 1.39 |
| establishment or maintenance of apical/basal cell polarity | 32 | 1.32 | 1.09 | 1.43 | 1.03 | 1.37 | 1.42 | 1.25 |
| establishment of apical/basal cell polarity | 8 | 1.65 | 1.12 | 1.18 | -1.06 | 1.49 | 1.45 | 1.45 |
| maintenance of apical/basal cell polarity | 7 | 1.32 | 1.27 | 1.59 | 1.12 | 1.39 | 1.26 | 1.25 |
| phosphatidylinositol binding | 27 | 1.13 | 1.11 | 1.46 | 1.09 | 1.31 | 1.15 | 1.26 |
| sperm chromatin condensation | 2 | 1.06 | 1.0 | -1.08 | -1.02 | -1.08 | -1.04 | -1.06 |
| spermatogenesis, exchange of chromosomal proteins | 1 | -1.14 | -1.02 | -1.18 | -1.04 | -1.12 | -1.15 | -1.23 |
| response to nicotine | 5 | -1.06 | -1.07 | -1.59 | -1.13 | 1.17 | 1.09 | 1.28 |
| behavioral response to nicotine | 1 | 1.09 | 1.23 | 3.2 | 1.05 | 2.53 | 1.91 | 1.95 |
| larval midgut cell programmed cell death | 3 | 2.11 | 1.08 | -1.08 | 1.13 | -1.02 | 1.36 | 1.42 |
| histone methyltransferase complex | 16 | 1.51 | 1.16 | 1.4 | 1.13 | 1.36 | 1.38 | 1.21 |
| ESC/E(Z) complex | 6 | 1.41 | 1.17 | 1.31 | 1.05 | 1.17 | 1.23 | 1.16 |
| hemocyte migration | 16 | -1.09 | 1.08 | 1.4 | 1.01 | 1.37 | 1.08 | 1.3 |
| ecdysone binding | 1 | 2.23 | 1.01 | -1.05 | 1.07 | 1.28 | -1.09 | 1.2 |
| FACT complex | 2 | -1.26 | 1.35 | 1.06 | 1.07 | 1.01 | -1.19 | 1.27 |
| PRC1 complex | 10 | 1.56 | 1.15 | 1.36 | 1.07 | 1.32 | 1.49 | 1.36 |
| sterol regulatory element binding protein cleavage | 4 | 1.66 | 1.17 | 1.17 | 1.07 | 1.36 | 1.15 | 1.2 |
| positive regulation of transcription via sterol regulatory element binding | 1 | 1.18 | 1.17 | 1.43 | 1.18 | 1.36 | -1.03 | 1.42 |
| appendage morphogenesis | 236 | 1.25 | 1.06 | 1.22 | 1.01 | 1.29 | 1.34 | 1.3 |
| limb morphogenesis | 54 | -1.01 | 1.08 | 1.1 | 1.01 | 1.36 | 1.25 | 1.25 |
| imaginal disc-derived limb morphogenesis | 54 | -1.01 | 1.08 | 1.1 | 1.01 | 1.36 | 1.25 | 1.25 |
| genitalia morphogenesis | 15 | 1.33 | 1.08 | 1.32 | 1.05 | 1.19 | 1.03 | 1.22 |
| imaginal disc-derived appendage morphogenesis | 234 | 1.25 | 1.06 | 1.23 | 1.01 | 1.29 | 1.34 | 1.31 |
| post-embryonic appendage morphogenesis | 227 | 1.26 | 1.06 | 1.23 | 1.01 | 1.3 | 1.36 | 1.32 |
| post-embryonic genitalia morphogenesis | 13 | 1.4 | 1.09 | 1.4 | 1.06 | 1.26 | 1.05 | 1.3 |
| post-embryonic limb morphogenesis | 53 | -1.02 | 1.08 | 1.1 | 1.01 | 1.37 | 1.25 | 1.26 |
| exon-exon junction complex | 4 | 1.04 | 1.14 | -1.13 | 1.22 | 1.04 | -1.18 | -1.31 |
| tube fusion | 19 | -1.07 | 1.1 | 1.35 | 1.01 | 1.36 | 1.22 | 1.46 |
| branch fusion, open tracheal system | 19 | -1.07 | 1.1 | 1.35 | 1.01 | 1.36 | 1.22 | 1.46 |
| tube formation | 13 | 1.06 | 1.1 | 1.18 | -1.05 | 1.05 | 1.27 | 1.36 |
| lumen formation, open tracheal system | 7 | 1.09 | 1.3 | 1.35 | 1.04 | 1.16 | 1.14 | 1.27 |
| regulation of tube size | 46 | 2.31 | -1.17 | 1.1 | -1.22 | 1.13 | 2.15 | 1.48 |
| regulation of tube size, open tracheal system | 43 | 2.4 | -1.2 | 1.11 | -1.24 | 1.13 | 2.23 | 1.49 |
| regulation of tube architecture, open tracheal system | 66 | 1.86 | -1.09 | 1.16 | -1.16 | 1.17 | 1.89 | 1.38 |
| epithelial cell type specification, open tracheal system | 8 | 1.08 | 1.1 | 1.45 | 1.06 | 1.34 | -1.23 | 1.4 |
| terminal cell fate specification, open tracheal system | 2 | -1.05 | 1.29 | 1.38 | 1.19 | 1.27 | -1.18 | 1.03 |
| negative regulation of terminal cell fate specification, open tracheal system | 4 | -1.37 | 1.26 | 1.57 | 1.19 | 1.57 | 1.09 | 1.17 |
| fusion cell fate specification | 2 | -1.28 | -1.08 | 2.4 | -1.03 | 1.63 | -2.97 | 1.01 |
| negative regulation of fusion cell fate specification | 3 | 1.21 | 1.19 | 1.16 | 1.18 | 1.53 | 1.57 | 1.32 |
| regulation of tube diameter, open tracheal system | 9 | 2.29 | -1.02 | 1.09 | -1.22 | -1.05 | 2.19 | 1.55 |
| regulation of tube length, open tracheal system | 24 | 1.81 | -1.04 | 1.23 | -1.13 | 1.27 | 1.82 | 1.36 |
| maintenance of epithelial integrity, open tracheal system | 12 | 1.32 | -1.13 | 1.03 | -1.19 | 1.26 | 1.44 | 1.38 |
| imaginal disc lineage restriction | 11 | 1.07 | 1.0 | 1.1 | 1.09 | 1.33 | 1.19 | 1.21 |
| embryonic hemopoiesis | 23 | -1.0 | 1.08 | 1.34 | 1.06 | 1.31 | 1.07 | 1.27 |
| embryonic hemocyte differentiation | 4 | 1.2 | 1.09 | 1.17 | 1.11 | 1.18 | 1.01 | 1.15 |
| embryonic plasmatocyte differentiation | 2 | 1.05 | 1.01 | 1.06 | 1.01 | 1.02 | -1.02 | 1.01 |
| embryonic crystal cell differentiation | 4 | 1.2 | 1.09 | 1.17 | 1.11 | 1.18 | 1.01 | 1.15 |
| post-embryonic hemopoiesis | 11 | 1.28 | 1.08 | 1.31 | 1.05 | 1.27 | 1.44 | 1.7 |
| larval lymph gland hemopoiesis | 11 | 1.28 | 1.08 | 1.31 | 1.05 | 1.27 | 1.44 | 1.7 |
| larval lymph gland hemocyte differentiation | 9 | 1.36 | 1.08 | 1.37 | 1.04 | 1.33 | 1.5 | 1.66 |
| lymph gland plasmatocyte differentiation | 1 | 1.08 | 1.07 | 1.03 | 1.0 | -1.04 | -1.0 | -1.04 |
| lymph gland crystal cell differentiation | 4 | 1.1 | 1.01 | 1.11 | 1.06 | 1.31 | 1.74 | 2.25 |
| lamellocyte differentiation | 4 | 1.23 | 1.27 | 1.61 | 1.19 | 1.65 | 1.45 | 1.54 |
| hemocyte proliferation | 10 | 1.23 | 1.17 | 1.5 | 1.16 | 1.47 | 1.44 | 1.37 |
| histone kinase activity | 3 | 1.34 | 1.23 | 1.39 | 1.12 | 1.46 | 1.54 | 1.4 |
| histone serine kinase activity | 2 | 1.43 | 1.39 | 1.53 | 1.13 | 1.4 | 1.89 | 1.58 |
| histone kinase activity (H3-S10 specific) | 1 | 1.82 | 1.38 | 1.39 | 1.27 | 1.17 | 2.09 | 1.46 |
| social behavior | 1 | -12.04 | -1.69 | 9.34 | -2.59 | -11.09 | -12.8 | -6.11 |
| larval foraging behavior | 1 | -12.04 | -1.69 | 9.34 | -2.59 | -11.09 | -12.8 | -6.11 |
| larval wandering behavior | 2 | -1.11 | 1.48 | 1.63 | -1.09 | 1.06 | -1.13 | 1.6 |
| female germline ring canal outer rim | 1 | 2.22 | 1.2 | -1.59 | -1.27 | 1.13 | 5.56 | 1.6 |
| female germline ring canal inner rim | 2 | 2.33 | 1.09 | -1.29 | -1.15 | -1.06 | 2.19 | 1.56 |
| histone threonine kinase activity | 1 | 1.19 | -1.04 | 1.15 | 1.11 | 1.6 | 1.03 | 1.09 |
| preblastoderm mitotic cell cycle | 2 | 6.0 | -2.29 | -2.45 | -2.24 | -2.05 | -1.18 | 2.53 |
| syncytial blastoderm mitotic cell cycle | 12 | 1.27 | 1.1 | 1.43 | 1.06 | 1.46 | 1.29 | 1.37 |
| hatching behavior | 2 | -5.42 | -1.46 | -1.97 | 1.51 | -1.45 | -12.25 | -3.05 |
| hatching | 3 | -2.59 | -1.22 | -1.47 | 1.31 | -1.21 | -4.45 | -1.84 |
| Rb-E2F complex | 5 | 1.18 | 1.3 | 1.41 | 1.2 | 1.32 | 1.41 | 1.26 |
| syncytial nuclear migration | 6 | 1.4 | 1.18 | 1.66 | 1.17 | 1.63 | 1.47 | 1.43 |
| nuclear axial expansion | 3 | 1.16 | 1.2 | 2.08 | 1.13 | 1.79 | 1.3 | 1.53 |
| larval central nervous system remodeling | 7 | 1.02 | -1.0 | 1.26 | -1.02 | 1.21 | -1.11 | 1.24 |
| posttranscriptional gene silencing by RNA | 37 | 1.12 | 1.04 | 1.18 | 1.1 | 1.27 | 1.33 | 1.07 |
| gene silencing by miRNA | 19 | 1.25 | 1.18 | 1.35 | 1.1 | 1.27 | 1.48 | 1.29 |
| siRNA binding | 4 | -1.07 | -1.68 | -1.17 | -1.84 | -1.07 | 1.05 | 2.69 |
| salt aversion | 1 | 1.12 | 1.06 | 1.12 | -1.01 | 1.01 | 1.05 | -1.06 |
| tracheal pit formation in open tracheal system | 4 | 1.93 | -1.11 | -1.01 | -1.1 | 1.21 | 1.33 | 1.64 |
| regulation of lamellocyte differentiation | 4 | 1.31 | 1.15 | -1.09 | -1.04 | 1.03 | 1.64 | 1.15 |
| negative regulation of lamellocyte differentiation | 3 | 1.53 | 1.17 | -1.05 | -1.05 | 1.11 | 2.06 | 1.23 |
| regulation of hemocyte proliferation | 3 | 1.68 | 1.14 | 1.31 | -1.06 | 1.35 | 1.48 | 1.49 |
| negative regulation of hemocyte proliferation | 2 | 1.12 | 1.36 | 1.4 | 1.12 | 1.42 | 1.34 | 1.46 |
| pupal development | 6 | 2.46 | -1.28 | -1.05 | -1.42 | -1.1 | 2.95 | -1.08 |
| prepupal development | 9 | 1.36 | 1.03 | 1.2 | -1.03 | 1.16 | 2.15 | 1.14 |
| spermathecum morphogenesis | 3 | -1.12 | 1.12 | -1.01 | 1.14 | 1.01 | 1.1 | 1.03 |
| cell competition in a multicellular organism | 8 | -1.44 | 1.26 | 1.72 | 1.12 | 1.65 | 1.02 | 1.13 |
| clypeo-labral disc development | 6 | 1.03 | 1.0 | 1.19 | 1.01 | 1.08 | 1.14 | 1.54 |
| eye-antennal disc development | 54 | 1.23 | 1.06 | 1.23 | 1.07 | 1.21 | 1.28 | 1.16 |
| genital disc development | 42 | 1.3 | -1.04 | 1.13 | 1.03 | 1.23 | 1.17 | 1.1 |
| haltere disc development | 10 | 1.31 | -1.02 | -1.01 | 1.12 | 1.13 | -1.17 | 1.04 |
| labial disc development | 4 | 1.69 | -1.22 | 1.25 | -1.11 | 1.22 | 1.33 | 1.1 |
| leg disc development | 68 | 1.06 | 1.07 | 1.11 | 1.01 | 1.33 | 1.23 | 1.24 |
| wing disc development | 298 | 1.28 | 1.06 | 1.23 | 1.02 | 1.24 | 1.31 | 1.31 |
| genital disc pattern formation | 8 | -1.0 | -1.63 | -1.11 | 1.02 | 1.25 | 1.06 | -1.49 |
| wing disc pattern formation | 64 | 1.33 | 1.02 | 1.11 | 1.04 | 1.23 | 1.29 | 1.24 |
| leg disc pattern formation | 17 | 1.41 | 1.01 | 1.1 | 1.07 | 1.05 | 1.04 | 1.19 |
| genital disc anterior/posterior pattern formation | 8 | -1.0 | -1.63 | -1.11 | 1.02 | 1.25 | 1.06 | -1.49 |
| determination of genital disc primordium | 10 | 1.07 | 1.03 | 1.07 | -1.02 | 1.18 | 1.04 | 1.37 |
| glutamate-cysteine ligase catalytic subunit binding | 1 | -1.69 | 1.43 | -1.23 | 1.48 | -1.02 | -3.82 | -1.18 |
| regulation of glutamate-cysteine ligase activity | 1 | -1.69 | 1.43 | -1.23 | 1.48 | -1.02 | -3.82 | -1.18 |
| positive regulation of glutamate-cysteine ligase activity | 1 | -1.69 | 1.43 | -1.23 | 1.48 | -1.02 | -3.82 | -1.18 |
| cytoneme assembly | 2 | -1.85 | -1.12 | 1.68 | 1.29 | 1.66 | 1.71 | -2.53 |
| germ cell attraction | 1 | -1.72 | -1.51 | 1.07 | 1.74 | 1.86 | -2.25 | -3.98 |
| germ cell repulsion | 2 | 1.22 | 1.02 | 1.17 | 1.16 | 2.02 | -1.19 | -1.06 |
| germ cell programmed cell death | 6 | 1.26 | 1.15 | 1.12 | 1.12 | 1.68 | 1.26 | 1.1 |
| ionotropic glutamate receptor signaling pathway | 2 | -1.26 | 1.06 | -1.07 | 1.01 | -1.06 | -1.06 | 1.04 |
| proctolin receptor activity | 1 | -1.08 | -1.05 | -1.14 | 1.14 | -1.06 | -1.04 | 1.03 |
| corazonin receptor activity | 1 | 1.24 | 1.25 | 1.2 | 1.13 | 1.08 | -1.04 | 1.04 |
| vitamin A biosynthetic process | 1 | 1.12 | -1.01 | 1.14 | 1.06 | 1.09 | 1.14 | -1.06 |
| tube morphogenesis | 104 | 1.21 | 1.02 | 1.27 | 1.0 | 1.31 | 1.36 | 1.2 |
| protein-arginine omega-N monomethyltransferase activity | 2 | 1.08 | 1.15 | 1.55 | 1.17 | 1.64 | 1.08 | 1.14 |
| protein-arginine omega-N asymmetric methyltransferase activity | 2 | 1.08 | 1.15 | 1.55 | 1.17 | 1.64 | 1.08 | 1.14 |
| peptidyl-arginine N-methylation | 3 | 1.19 | 1.12 | 1.45 | 1.13 | 1.36 | 1.2 | 1.09 |
| peptidyl-arginine omega-N-methylation | 3 | 1.19 | 1.12 | 1.45 | 1.13 | 1.36 | 1.2 | 1.09 |
| alpha-1,4-N-acetylgalactosaminyltransferase activity | 1 | 3.56 | 1.12 | 1.2 | 1.23 | 1.49 | 1.56 | 1.46 |
| synaptic transmission, glutamatergic | 2 | -1.04 | 1.09 | 1.07 | 1.06 | 1.14 | 1.17 | 1.1 |
| UDP-galactosyltransferase activity | 13 | -1.53 | 1.04 | 1.17 | 1.15 | 1.2 | -1.26 | -1.01 |
| UDP-glucosyltransferase activity | 10 | 1.76 | -1.24 | -1.02 | -1.41 | 1.08 | 1.69 | 1.09 |
| UDP-xylosyltransferase activity | 1 | 1.17 | 1.3 | 1.39 | 1.35 | 1.31 | 1.22 | 1.24 |
| ciliary rootlet | 1 | 1.02 | 1.34 | 1.17 | 1.22 | -1.17 | 1.04 | 1.06 |
| nuclear hormone receptor binding | 4 | 1.17 | 1.22 | 1.31 | 1.27 | 1.44 | 1.44 | 1.01 |
| steroid hormone receptor binding | 2 | 1.48 | 1.23 | 1.51 | 1.28 | 1.77 | 1.32 | 1.19 |
| external genitalia morphogenesis | 1 | -1.17 | -1.03 | -1.15 | -1.16 | -1.43 | -1.09 | -1.47 |
| gonad morphogenesis | 4 | -1.42 | 1.18 | 1.32 | 1.12 | 1.16 | -1.2 | 1.04 |
| genital disc sexually dimorphic development | 5 | 1.78 | 1.05 | 1.16 | -1.11 | -1.05 | -1.07 | 1.31 |
| multicellular organism growth | 3 | 1.09 | 1.05 | 1.05 | -1.19 | -1.03 | 1.35 | 1.92 |
| organ growth | 18 | 1.41 | 1.18 | 1.37 | 1.13 | 1.39 | 1.29 | 1.29 |
| NuA4 histone acetyltransferase complex | 5 | 1.44 | 1.1 | 1.23 | 1.2 | 1.27 | 1.53 | -1.03 |
| protein mannosylation | 2 | 1.57 | 1.01 | 1.07 | -1.18 | -1.11 | 1.61 | 1.23 |
| protein O-linked mannosylation | 2 | 1.57 | 1.01 | 1.07 | -1.18 | -1.11 | 1.61 | 1.23 |
| endocrine system development | 5 | 1.34 | 1.03 | 1.09 | -1.08 | -1.01 | 1.09 | 1.11 |
| ring gland development | 5 | 1.34 | 1.03 | 1.09 | -1.08 | -1.01 | 1.09 | 1.11 |
| exocrine system development | 141 | 1.08 | 1.0 | 1.15 | 1.05 | 1.27 | 1.18 | 1.17 |
| phthalate binding | 1 | 1.12 | 1.0 | 1.13 | 1.06 | 1.1 | -1.01 | -1.1 |
| diphenyl phthalate binding | 1 | 1.12 | 1.0 | 1.13 | 1.06 | 1.1 | -1.01 | -1.1 |
| dibutyl phthalate binding | 1 | 1.12 | 1.0 | 1.13 | 1.06 | 1.1 | -1.01 | -1.1 |
| ethanol binding | 1 | 1.12 | 1.0 | 1.13 | 1.06 | 1.1 | -1.01 | -1.1 |
| spiracle morphogenesis, open tracheal system | 16 | 1.23 | 1.0 | 1.11 | 1.01 | 1.19 | 1.45 | 1.19 |
| miRNA loading onto RISC involved in gene silencing by miRNA | 1 | 1.78 | 1.14 | -1.44 | 1.08 | 1.0 | 3.89 | 1.83 |
| segmentation | 206 | 1.14 | 1.07 | 1.24 | 1.03 | 1.25 | 1.25 | 1.21 |
| central nervous system segmentation | 4 | -1.45 | -1.33 | 2.24 | -1.39 | -1.52 | -1.44 | -1.38 |
| brain segmentation | 4 | -1.45 | -1.33 | 2.24 | -1.39 | -1.52 | -1.44 | -1.38 |
| appendage segmentation | 20 | -1.29 | 1.07 | -1.1 | -1.14 | 1.65 | 1.34 | 1.29 |
| imaginal disc-derived leg segmentation | 20 | -1.29 | 1.07 | -1.1 | -1.14 | 1.65 | 1.34 | 1.29 |
| head segmentation | 22 | 1.16 | -1.02 | 1.07 | 1.04 | -1.04 | -1.02 | 1.02 |
| anterior head segmentation | 9 | 1.09 | -1.1 | 1.06 | 1.06 | -1.02 | -1.13 | -1.01 |
| posterior head segmentation | 15 | 1.05 | -1.01 | 1.05 | -1.0 | 1.02 | 1.0 | 1.07 |
| trunk segmentation | 15 | 1.21 | -1.04 | -1.03 | 1.0 | -1.02 | 1.0 | 1.11 |
| specification of segmental identity, intercalary segment | 1 | -1.22 | 1.08 | -1.24 | -1.01 | -1.23 | -1.21 | -1.07 |
| specification of segmental identity, trunk | 5 | 1.18 | 1.16 | 1.14 | 1.3 | -1.04 | 1.14 | -1.03 |
| chitin-based larval cuticle pattern formation | 9 | -1.18 | 1.14 | 1.32 | 1.06 | 1.18 | -1.12 | 1.22 |
| tube development | 132 | 1.13 | 1.03 | 1.25 | 1.01 | 1.26 | 1.33 | 1.19 |
| regulation of tube diameter | 12 | 2.01 | 1.03 | 1.06 | -1.16 | 1.02 | 1.94 | 1.48 |
| regulation of Malpighian tubule diameter | 3 | 1.35 | 1.21 | -1.03 | 1.01 | 1.25 | 1.33 | 1.29 |
| regulation of Malpighian tubule size | 4 | 1.53 | 1.16 | 1.13 | -1.02 | 1.37 | 1.52 | 1.41 |
| inositol pentakisphosphate 2-kinase activity | 1 | -1.11 | 1.15 | 1.19 | 1.05 | 1.03 | 1.14 | 1.11 |
| Hedgehog signaling complex | 4 | 2.07 | 1.18 | 1.21 | 1.01 | 1.19 | 2.13 | 1.37 |
| ecdysteroid 25-hydroxylase activity | 1 | 1.02 | -1.19 | -1.05 | 1.18 | -1.22 | -1.04 | -1.19 |
| regulation of dephosphorylation | 8 | 1.15 | 1.12 | 1.06 | 1.12 | 1.09 | 1.13 | 1.04 |
| regulation of protein dephosphorylation | 6 | 1.25 | 1.15 | 1.13 | 1.13 | 1.17 | 1.25 | 1.12 |
| negative regulation of dephosphorylation | 4 | 1.52 | 1.1 | 1.03 | 1.13 | 1.22 | 1.28 | 1.01 |
| negative regulation of protein dephosphorylation | 4 | 1.52 | 1.1 | 1.03 | 1.13 | 1.22 | 1.28 | 1.01 |
| wing and notum subfield formation | 12 | 1.93 | -1.1 | 1.05 | -1.1 | 1.08 | 1.14 | 1.87 |
| notum cell fate specification | 4 | 1.88 | -1.36 | -1.26 | -1.21 | 1.01 | 1.16 | 3.54 |
| wing cell fate specification | 3 | -1.07 | 1.05 | 1.06 | -1.14 | 1.2 | -1.05 | 2.07 |
| wound healing, spreading of epidermal cells | 1 | -1.04 | -1.12 | 1.46 | 1.28 | 1.4 | -1.15 | 1.1 |
| scab formation | 1 | 1.24 | 1.07 | 1.06 | 1.19 | 1.06 | 1.05 | 1.04 |
| hair cell differentiation | 41 | 1.43 | 1.05 | 1.43 | -1.01 | 1.36 | 1.42 | 1.36 |
| non-sensory hair organization | 41 | 1.43 | 1.05 | 1.43 | -1.01 | 1.36 | 1.42 | 1.36 |
| imaginal disc-derived wing hair organization | 40 | 1.38 | 1.06 | 1.45 | -1.0 | 1.37 | 1.37 | 1.36 |
| imaginal disc-derived wing hair outgrowth | 1 | -2.13 | 1.07 | 2.17 | -1.03 | 1.35 | -1.42 | -1.02 |
| imaginal disc-derived wing hair site selection | 3 | 1.72 | -1.01 | 1.03 | -1.08 | 1.97 | 1.34 | 1.24 |
| maintenance of imaginal disc-derived wing hair orientation | 3 | 1.52 | 1.13 | 1.79 | 1.02 | 1.81 | 2.37 | 1.98 |
| male germline ring canal | 3 | -1.27 | 1.18 | 1.18 | 1.26 | 1.55 | -1.01 | 1.01 |
| female germline ring canal | 3 | 1.66 | 1.08 | -1.16 | 1.04 | 1.18 | 1.57 | 1.3 |
| enhancer binding | 18 | 1.2 | 1.05 | 1.07 | -1.02 | 1.0 | 1.04 | 1.19 |
| transcriptionally active chromatin | 19 | 1.06 | 1.16 | 1.2 | 1.2 | 1.33 | 1.29 | 1.09 |
| transcriptionally silent chromatin | 1 | 1.82 | 1.17 | 1.65 | -1.11 | 1.49 | 1.95 | 1.38 |
| hippo signaling cascade | 7 | 1.44 | 1.16 | 1.3 | 1.05 | 1.29 | 1.47 | 1.15 |
| regulation of hippo signaling cascade | 2 | 3.69 | 1.25 | 1.96 | -1.14 | 1.8 | 2.24 | 2.45 |
| negative regulation of hippo signaling cascade | 1 | 3.54 | 1.08 | 1.48 | -1.26 | 1.42 | 1.25 | 1.69 |
| fatty-acyl-CoA metabolic process | 4 | 1.86 | 1.19 | 1.27 | 1.15 | 1.54 | 1.19 | 1.01 |
| histone locus body | 7 | 1.03 | 1.04 | 1.16 | 1.17 | 1.27 | 1.05 | 1.03 |
| microtubule plus end | 2 | 1.59 | 1.18 | 1.94 | -1.04 | 1.66 | 1.95 | 1.8 |
| thioester metabolic process | 6 | 2.01 | 1.03 | 1.27 | 1.12 | 1.42 | 1.33 | 1.06 |
| thioester biosynthetic process | 5 | 2.3 | 1.04 | 1.28 | 1.15 | 1.53 | 1.22 | 1.09 |
| Roundabout signaling pathway | 2 | 1.81 | 1.11 | 1.52 | 1.11 | 1.43 | 1.62 | 1.25 |
| regulation of Roundabout signaling pathway | 1 | 1.04 | 1.02 | -1.06 | 1.23 | 1.3 | 3.59 | 1.24 |
| histone-serine phosphorylation | 1 | 1.24 | 1.29 | 1.32 | 1.24 | 1.1 | -1.34 | -1.14 |
| regulation of mitotic prometaphase | 1 | 1.18 | -1.09 | 1.06 | 1.12 | 1.44 | -1.15 | -1.22 |
| protein localization to synapse | 1 | 1.3 | 1.23 | 1.1 | 1.08 | 1.16 | 2.99 | 1.24 |
| copper ion transmembrane transport | 4 | -1.26 | 1.17 | 1.07 | 1.07 | -1.01 | -1.03 | 1.07 |
| phosphate ion transmembrane transport | 4 | -1.25 | -1.17 | -1.77 | -1.14 | -1.01 | 2.78 | -1.28 |
| maintenance of protein localization in endoplasmic reticulum | 3 | -1.9 | 1.28 | -1.09 | 1.08 | -1.19 | -2.08 | 1.34 |
| regulation of chromatin binding | 3 | -1.0 | 1.07 | 1.16 | 1.27 | 1.61 | -1.41 | -1.14 |
| non-canonical Wnt receptor signaling pathway | 6 | 1.09 | -1.01 | 1.23 | -1.1 | 1.27 | 1.47 | 1.72 |
| purinergic receptor activity | 1 | -1.32 | 1.16 | 1.21 | -1.14 | -1.21 | 1.47 | 1.08 |
| signaling adaptor activity | 10 | 1.08 | 1.02 | 1.26 | 1.08 | 1.17 | 1.33 | -1.04 |
| establishment of protein localization in extracellular region | 1 | 1.79 | 1.14 | -1.18 | 1.07 | 1.25 | -5.71 | 1.37 |
| positive regulation of Wnt receptor signaling pathway by establishment of Wnt protein localization in extracellular region | 1 | 1.79 | 1.14 | -1.18 | 1.07 | 1.25 | -5.71 | 1.37 |
| protein deacylation | 11 | 1.53 | 1.15 | 1.12 | 1.23 | 1.27 | 1.36 | 1.13 |
| RNA stem-loop binding | 2 | 1.54 | -1.06 | 1.11 | 1.22 | 1.17 | 1.23 | -1.02 |
| snRNA stem-loop binding | 1 | 1.41 | -1.11 | 1.3 | 1.34 | 1.31 | 1.54 | -1.14 |
| ceramide transporter activity | 1 | -2.39 | 1.47 | 1.51 | 1.07 | -1.25 | -2.16 | 1.24 |
| ER to Golgi ceramide transport | 1 | -2.39 | 1.47 | 1.51 | 1.07 | -1.25 | -2.16 | 1.24 |
| juvenile hormone mediated signaling pathway | 3 | -1.19 | 1.31 | 1.42 | 1.17 | 1.93 | 1.0 | 1.17 |
| ceramide transport | 1 | -2.39 | 1.47 | 1.51 | 1.07 | -1.25 | -2.16 | 1.24 |
| maintenance of blood-brain barrier | 1 | 7.29 | -3.52 | -1.24 | -1.54 | -1.02 | 7.02 | -1.25 |
| multicellular organismal signaling | 223 | 1.08 | 1.07 | 1.2 | 1.03 | 1.19 | 1.22 | 1.13 |
| signal maturation | 3 | -1.03 | 1.15 | 1.23 | 1.08 | 1.5 | -1.16 | 1.1 |
| purine ribonucleoside triphosphate binding | 806 | 1.08 | 1.1 | 1.19 | 1.08 | 1.22 | 1.17 | 1.09 |
| exploration behavior | 1 | 3.78 | 1.32 | 1.35 | -1.13 | 1.11 | 1.73 | 1.58 |
| locomotory exploration behavior | 1 | 3.78 | 1.32 | 1.35 | -1.13 | 1.11 | 1.73 | 1.58 |
| oligopeptide transmembrane transporter activity | 3 | 1.69 | -1.17 | -1.51 | -1.15 | -1.07 | 1.17 | 1.2 |
| cell proliferation involved in compound eye morphogenesis | 1 | -1.2 | 1.15 | 1.53 | 1.19 | 1.27 | 1.15 | 1.24 |
| ribonucleoprotein granule | 26 | 1.39 | 1.14 | 1.33 | 1.09 | 1.3 | 1.57 | 1.15 |
| ubiquitin-specific protease activator activity | 1 | 1.14 | 1.09 | 1.34 | -1.05 | 1.24 | 1.35 | 2.12 |
| egg coat formation | 12 | 1.14 | 1.01 | -1.05 | -1.02 | -1.08 | 1.01 | -1.0 |
| structural constituent of egg coat | 5 | -1.04 | 1.0 | -1.05 | -1.03 | -1.16 | -1.05 | -1.08 |
| egg coat | 5 | -1.16 | -1.01 | -1.07 | -1.03 | -1.18 | -1.17 | -1.07 |
| reciprocal DNA recombination | 22 | 1.06 | 1.12 | 1.07 | 1.13 | 1.17 | 1.01 | 1.01 |
| ciliary transition zone | 2 | -1.07 | -1.03 | -1.02 | 1.13 | -1.1 | -1.09 | -1.15 |
| skeletal muscle cell differentiation | 27 | 1.01 | 1.06 | 1.27 | -1.02 | 1.21 | 1.27 | 1.14 |
| response to topologically incorrect protein | 2 | 1.04 | 1.38 | 1.16 | -1.1 | 1.05 | 1.03 | 1.56 |
| cellular response to topologically incorrect protein | 1 | -1.05 | 1.27 | -1.17 | -1.24 | -1.49 | -1.01 | 1.19 |
| peptidyl-threonine dephosphorylation | 1 | 1.22 | 1.23 | -1.22 | 1.31 | 1.05 | -1.8 | -1.36 |
| endodermal cell differentiation | 1 | 8.12 | -1.58 | 1.44 | -1.54 | 1.01 | 1.69 | 1.21 |
| pre-mRNA binding | 1 | 1.28 | 1.07 | 1.73 | 1.12 | 1.48 | 1.94 | 1.41 |
| limb joint morphogenesis | 8 | -2.19 | 1.03 | -1.69 | -1.64 | 2.1 | 1.1 | 1.32 |
| filtration diaphragm | 3 | -3.44 | 1.01 | 1.01 | 1.15 | 1.01 | -2.84 | -1.29 |
| filtration diaphragm assembly | 2 | -1.2 | -1.04 | -1.19 | 1.02 | -1.08 | -1.21 | -1.15 |
| nephrocyte diaphragm assembly | 2 | -1.2 | -1.04 | -1.19 | 1.02 | -1.08 | -1.21 | -1.15 |
| muscle cell chemotaxis toward tendon cell | 1 | 1.49 | 1.07 | 1.15 | 1.04 | -1.03 | 1.53 | -1.09 |
| presynaptic periactive zone | 2 | 1.13 | 1.16 | 1.42 | 1.03 | 1.54 | 1.68 | 1.68 |
| acroblast | 1 | -1.29 | 1.49 | 1.22 | 1.13 | 1.05 | 1.04 | 1.36 |
| fucosylation | 2 | -1.26 | 1.19 | -1.04 | 1.47 | 1.01 | -1.41 | -1.39 |
| protein O-linked fucosylation | 2 | -1.26 | 1.19 | -1.04 | 1.47 | 1.01 | -1.41 | -1.39 |
| N-glycan fucosylation | 1 | -1.64 | 1.12 | -1.15 | 1.5 | 1.42 | -1.98 | -1.63 |
| minus-end specific microtubule depolymerization | 2 | -1.34 | 1.21 | 1.12 | 1.01 | 1.15 | 1.13 | 1.12 |
| purine nucleotide-sugar transport | 2 | -1.26 | 1.19 | -1.04 | 1.47 | 1.01 | -1.41 | -1.39 |
| extracellular ammonia-gated ion channel activity | 1 | -1.06 | -1.02 | 1.01 | 1.08 | -1.26 | -1.1 | -1.15 |
| extracellular phenylacetaldehyde-gated ion channel activity | 1 | -1.38 | 1.0 | -1.11 | -1.02 | -1.17 | -1.19 | 1.01 |
| GDP-fucose import into endoplasmic reticulum lumen | 1 | 1.04 | 1.26 | 1.06 | 1.45 | -1.4 | -1.0 | -1.18 |
| GDP-fucose import into Golgi lumen | 1 | -1.64 | 1.12 | -1.15 | 1.5 | 1.42 | -1.98 | -1.63 |
| small molecule binding | 1251 | 1.1 | 1.05 | 1.07 | 1.05 | 1.14 | 1.14 | 1.07 |
| fatty acid beta-oxidation multienzyme complex | 2 | 2.42 | 1.07 | -1.08 | 1.02 | 1.26 | 1.66 | -1.08 |
| kynurenine aminotransferase activity | 1 | 1.14 | 1.23 | -1.25 | 1.13 | -1.05 | 1.32 | 1.04 |
| epidermal growth factor receptor ligand maturation | 2 | -1.08 | 1.07 | 1.03 | -1.09 | 1.34 | -1.21 | 1.17 |
| peptide bond cleavage involved in epidermal growth factor receptor ligand maturation | 1 | -1.2 | -1.0 | -1.26 | -1.23 | 1.12 | -1.78 | 1.11 |
| netrin-activated signaling pathway | 1 | -10.89 | 1.57 | 1.23 | -1.08 | -1.19 | -6.39 | 1.44 |
| signaling receptor activity | 360 | 1.11 | -1.02 | 1.05 | -1.02 | 1.03 | 1.11 | 1.05 |
| termination of G-protein coupled receptor signaling pathway | 9 | 1.61 | 1.14 | 1.6 | 1.12 | 1.38 | 1.47 | 1.34 |
| negative regulation of BMP signaling pathway by negative regulation of BMP secretion | 1 | -1.14 | 1.11 | -1.09 | 1.17 | -1.12 | -1.22 | -1.05 |
| ERBB signaling pathway | 34 | 1.44 | 1.08 | 1.27 | 1.08 | 1.29 | 1.29 | 1.22 |
| establishment of mitotic spindle localization | 9 | -1.21 | 1.28 | 1.27 | 1.13 | 1.32 | 1.06 | 1.1 |
| chitin-based cuticle development | 75 | 1.22 | -1.06 | -1.0 | -1.08 | -1.04 | 1.24 | 1.13 |
| chitin-based cuticle attachment to epithelium | 3 | 6.48 | -1.33 | -1.2 | -1.65 | -1.09 | 5.63 | 1.91 |
| protein-based cuticle attachment to epithelium | 12 | 1.66 | -1.21 | -1.1 | -1.18 | -1.09 | 1.51 | 1.5 |
| regulation of growth rate | 1 | 1.08 | -1.16 | -1.09 | 1.12 | -1.12 | 1.04 | -1.07 |
| locomotion | 406 | 1.12 | 1.06 | 1.21 | 1.02 | 1.26 | 1.23 | 1.22 |
| regulation of locomotion | 16 | 1.62 | -1.06 | 1.25 | -1.04 | 1.18 | 1.21 | 1.39 |
| negative regulation of locomotion | 2 | 2.1 | 1.1 | 1.42 | -1.08 | 1.39 | 1.72 | 1.26 |
| regulation of multicellular organism growth | 30 | 1.09 | 1.12 | 1.29 | 1.0 | 1.23 | 1.41 | 1.42 |
| negative regulation of multicellular organism growth | 4 | 1.6 | 1.06 | 1.17 | -1.17 | 1.27 | 2.71 | 1.48 |
| embryonic cleavage | 4 | 1.73 | 1.15 | 1.27 | 1.05 | 1.09 | 1.55 | 1.23 |
| positive regulation of multicellular organism growth | 18 | -1.05 | 1.16 | 1.29 | 1.01 | 1.23 | 1.35 | 1.42 |
| regulation of meiosis | 6 | 1.06 | 1.25 | 1.33 | 1.14 | 1.41 | 1.2 | 1.19 |
| establishment of nucleus localization | 24 | 1.13 | 1.16 | 1.28 | 1.13 | 1.3 | 1.19 | 1.15 |
| regulation of gene expression, epigenetic | 101 | 1.25 | 1.1 | 1.27 | 1.1 | 1.29 | 1.34 | 1.18 |
| snRNA modification | 2 | -1.23 | 1.23 | 1.84 | 1.26 | 1.56 | -1.27 | 1.05 |
| regulation of development, heterochronic | 11 | 1.25 | 1.11 | 1.24 | 1.06 | 1.18 | 1.2 | 1.12 |
| regulation of fibroblast growth factor receptor signaling pathway | 2 | -1.9 | 1.49 | 1.19 | 1.1 | 1.08 | -1.77 | 1.19 |
| negative regulation of fibroblast growth factor receptor signaling pathway | 2 | -1.9 | 1.49 | 1.19 | 1.1 | 1.08 | -1.77 | 1.19 |
| thermosensory behavior | 6 | 1.6 | -1.4 | -1.4 | -1.32 | -1.22 | 1.3 | 1.62 |
| DNA endoreduplication | 16 | 1.23 | 1.15 | 1.24 | 1.15 | 1.47 | 1.49 | 1.25 |
| fluid transport | 13 | 1.15 | -1.04 | -1.12 | -1.03 | 1.05 | 1.11 | 1.09 |
| epithelial fluid transport | 9 | 1.44 | -1.07 | -1.0 | -1.13 | 1.2 | 1.6 | 1.1 |
| olfactory behavior | 83 | 1.02 | 1.05 | 1.18 | -1.02 | 1.16 | 1.12 | 1.18 |
| cellular acyl-CoA homeostasis | 6 | -3.11 | -1.44 | -2.62 | 1.15 | 1.36 | -3.71 | -1.6 |
| rhabdomere development | 25 | 1.09 | 1.1 | 1.42 | -1.0 | 1.23 | 1.32 | 1.22 |
| regulation of dopamine metabolic process | 2 | 1.15 | -1.11 | 1.14 | 1.21 | 1.14 | -1.17 | 1.36 |
| histone methyltransferase activity | 17 | 1.26 | 1.24 | 1.47 | 1.08 | 1.41 | 1.32 | 1.32 |
| regulation of epidermal growth factor receptor signaling pathway | 30 | 1.05 | 1.05 | 1.28 | 1.05 | 1.31 | 1.06 | 1.19 |
| negative regulation of epidermal growth factor receptor signaling pathway | 20 | 1.01 | 1.05 | 1.24 | 1.07 | 1.24 | 1.05 | 1.14 |
| wound healing | 24 | 1.08 | -1.07 | 1.03 | -1.16 | 1.04 | 1.25 | 1.33 |
| long-term strengthening of neuromuscular junction | 2 | -1.81 | 1.29 | 2.08 | -1.04 | 1.39 | 1.52 | 1.29 |
| gliogenesis | 57 | 1.43 | -1.08 | 1.21 | -1.07 | 1.26 | 1.62 | 1.2 |
| glial cell growth | 6 | -1.14 | -1.15 | -1.08 | -1.34 | 1.14 | 2.33 | 1.04 |
| perineurial glial growth | 6 | -1.14 | -1.15 | -1.08 | -1.34 | 1.14 | 2.33 | 1.04 |
| establishment of ommatidial planar polarity | 48 | 1.14 | 1.05 | 1.16 | -1.01 | 1.24 | 1.16 | 1.5 |
| regulation of catecholamine metabolic process | 3 | 1.05 | 1.06 | 1.07 | 1.15 | 1.1 | -1.22 | 1.43 |
| maintenance of oocyte nucleus location involved in oocyte dorsal/ventral axis specification | 1 | 1.08 | 1.36 | 1.49 | -1.03 | 1.39 | 1.74 | 1.47 |
| leucokinin receptor activity | 1 | 1.19 | -1.18 | -1.07 | -1.08 | 2.45 | 1.28 | 2.37 |
| intraflagellar transport | 4 | -1.17 | 1.08 | 1.02 | 1.06 | 1.03 | -1.03 | -1.17 |
| cell migration involved in gastrulation | 8 | 1.53 | -1.02 | 1.51 | -1.06 | 1.32 | 1.72 | 1.01 |
| germ-line stem cell division | 32 | 1.22 | 1.09 | 1.29 | 1.05 | 1.29 | 1.16 | 1.18 |
| 5,10-methylenetetrahydrofolate-dependent methyltransferase activity | 2 | 1.05 | 1.03 | -1.1 | 1.26 | 1.31 | 1.33 | -1.09 |
| macrophage activation | 1 | 2.18 | -2.11 | 2.19 | 1.49 | 1.48 | 1.37 | 1.38 |
| regulation of cell proliferation | 90 | 1.31 | 1.08 | 1.33 | 1.08 | 1.45 | 1.37 | 1.19 |
| fructose 1,6-bisphosphate 1-phosphatase activity | 1 | 6.67 | 1.2 | 1.32 | -2.53 | -1.76 | 2.31 | 2.31 |
| neurotransmitter metabolic process | 6 | -1.15 | 1.13 | -1.61 | -1.13 | -1.28 | -2.46 | 1.45 |
| neurotransmitter catabolic process | 2 | -1.99 | 1.48 | -3.26 | -1.27 | -2.14 | -11.86 | 3.38 |
| meiotic DNA double-strand break formation | 3 | 1.11 | 1.2 | 1.11 | 1.16 | 1.03 | 1.04 | -1.07 |
| early meiotic recombination nodule assembly | 1 | -1.21 | 1.44 | 1.1 | 1.2 | 1.03 | -1.18 | 1.17 |
| late meiotic recombination nodule assembly | 1 | -1.21 | 1.44 | 1.1 | 1.2 | 1.03 | -1.18 | 1.17 |
| retrograde transport, endosome to Golgi | 2 | -1.61 | 1.37 | 1.68 | 1.0 | 1.09 | 1.3 | 1.14 |
| lipoprotein metabolic process | 52 | -1.06 | 1.01 | 1.05 | 1.04 | 1.15 | -1.02 | -1.13 |
| lipoprotein biosynthetic process | 50 | -1.11 | 1.0 | 1.04 | 1.04 | 1.13 | -1.02 | -1.14 |
| lipoprotein catabolic process | 2 | 2.75 | 1.12 | 1.29 | 1.19 | 1.61 | 1.16 | 1.22 |
| neurotransmitter binding | 1 | -1.34 | -1.11 | -1.08 | -1.04 | -1.26 | -1.15 | -1.15 |
| acetylcholine binding | 1 | -1.34 | -1.11 | -1.08 | -1.04 | -1.26 | -1.15 | -1.15 |
| heme metabolic process | 9 | 1.28 | 1.21 | 1.24 | 1.17 | 1.12 | 1.19 | -1.04 |
| SH2 domain binding | 4 | 1.97 | -1.06 | 1.04 | -1.03 | 1.04 | 1.79 | 1.15 |
| lysophosphatidic acid acyltransferase activity | 1 | 1.37 | 1.22 | 1.4 | 1.12 | 1.37 | 1.84 | 1.71 |
| nuclear outer membrane-endoplasmic reticulum membrane network | 66 | -1.13 | 1.17 | 1.14 | 1.11 | 1.13 | -1.15 | 1.14 |
| regulation of protein catabolic process | 10 | 1.41 | 1.11 | 1.26 | 1.18 | 1.24 | 1.23 | 1.21 |
| negative regulation of protein catabolic process | 3 | 1.42 | 1.07 | 1.19 | 1.26 | 1.18 | -1.01 | -1.02 |
| xenobiotic catabolic process | 4 | -1.27 | -1.2 | -4.97 | -1.06 | -1.84 | -3.14 | 1.18 |
| cellular ketone metabolic process | 259 | 1.01 | -1.01 | -1.13 | -1.02 | 1.01 | 1.08 | -1.04 |
| ketone biosynthetic process | 12 | 1.31 | -1.17 | -1.03 | -1.07 | 1.06 | 1.52 | 1.13 |
| ketone catabolic process | 1 | 1.07 | -1.12 | 1.15 | -1.13 | -1.09 | 1.14 | -1.07 |
| terpene metabolic process | 1 | 1.12 | -1.01 | 1.14 | 1.06 | 1.09 | 1.14 | -1.06 |
| 1-aminocyclopropane-1-carboxylate biosynthetic process | 2 | 1.14 | 1.17 | -1.17 | -1.01 | 1.02 | 1.07 | -1.0 |
| response to cocaine | 13 | 1.28 | 1.17 | 1.57 | 1.07 | 1.39 | 1.83 | 1.27 |
| response to chemical stimulus | 654 | 1.11 | 1.04 | 1.13 | 1.0 | 1.14 | 1.18 | 1.15 |
| tissue regeneration | 8 | -1.03 | -1.05 | -1.06 | -1.23 | 1.0 | 1.47 | 1.44 |
| maintenance of polarity of follicular epithelium | 2 | -2.09 | 1.21 | 1.5 | 1.33 | 2.17 | -1.63 | -1.29 |
| establishment of planar polarity of embryonic epithelium | 1 | -2.32 | 1.45 | 1.45 | 1.19 | 1.32 | -1.02 | 1.2 |
| maintenance of polarity of embryonic epithelium | 1 | 1.75 | -1.09 | 1.21 | -1.06 | -1.12 | -1.14 | -1.06 |
| ribosome assembly | 3 | 1.33 | 1.18 | 1.17 | 1.23 | 1.06 | 1.03 | 1.04 |
| mature ribosome assembly | 1 | -1.17 | 1.17 | 1.23 | 1.32 | 1.08 | -1.28 | 1.0 |
| neuropeptide F receptor activity | 1 | -1.53 | -1.53 | 2.77 | -1.11 | -1.08 | -1.73 | 1.51 |
| ribosomal large subunit biogenesis | 1 | 1.48 | 1.28 | 1.8 | 1.26 | 1.7 | 1.35 | 1.33 |
| ribosomal small subunit biogenesis | 2 | 1.29 | 1.16 | 1.18 | 1.34 | 1.24 | 1.04 | 1.05 |
| peptide binding | 14 | -1.09 | 1.19 | -1.02 | 1.09 | -1.0 | -1.25 | 1.18 |
| host cell surface receptor binding | 1 | 2.63 | -1.41 | 1.69 | -1.89 | -1.48 | 14.61 | -1.02 |
| dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity | 1 | -1.04 | -1.0 | 1.06 | -1.08 | -1.02 | -1.02 | -1.17 |
| sphingolipid delta-4 desaturase activity | 1 | -6.62 | 1.41 | 1.16 | 1.17 | 1.23 | -3.29 | -1.25 |
| xylosyltransferase activity | 1 | 1.17 | 1.3 | 1.39 | 1.35 | 1.31 | 1.22 | 1.24 |
| structural constituent of cuticle | 133 | 1.15 | -1.12 | -1.18 | -1.14 | -1.24 | -1.07 | -1.1 |
| molting cycle | 55 | 1.16 | -1.03 | 1.01 | -1.09 | -1.04 | 1.19 | 1.17 |
| regulation of fatty acid biosynthetic process | 1 | 1.18 | 1.17 | 1.43 | 1.18 | 1.36 | -1.03 | 1.42 |
| specification of segmental identity, mandibular segment | 2 | -1.03 | 1.02 | 1.14 | 1.06 | -1.05 | -1.03 | 1.17 |
| regulation of protein import into nucleus | 30 | 1.19 | 1.1 | 1.26 | 1.1 | 1.28 | 1.31 | 1.24 |
| positive regulation of protein import into nucleus | 18 | 1.13 | 1.06 | 1.22 | 1.1 | 1.37 | 1.17 | 1.13 |
| negative regulation of protein import into nucleus | 5 | 2.19 | 1.12 | 1.26 | 1.06 | 1.21 | 2.05 | 1.7 |
| protein kinase C deactivation | 1 | -1.49 | 1.25 | 1.1 | 1.05 | -1.11 | -1.19 | 1.29 |
| penicillin metabolic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| penicillin biosynthetic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| regulation of phosphorylation | 73 | 1.32 | 1.12 | 1.35 | 1.09 | 1.31 | 1.39 | 1.22 |
| negative regulation of phosphorylation | 4 | 1.29 | -1.03 | 1.47 | -1.02 | 1.17 | 1.39 | 1.53 |
| positive regulation of phosphorylation | 8 | 1.51 | 1.21 | 1.82 | 1.05 | 1.45 | 2.01 | 1.56 |
| heparan sulfate N-acetylglucosaminyltransferase activity | 1 | 1.03 | 1.3 | 1.31 | 1.09 | 1.28 | -1.03 | 1.49 |
| taxis | 226 | 1.11 | 1.04 | 1.18 | -1.0 | 1.24 | 1.24 | 1.24 |
| phototaxis | 14 | 1.37 | -1.07 | 1.01 | -1.18 | -1.0 | 1.35 | 1.36 |
| gravitaxis | 12 | 1.09 | -1.01 | 1.03 | 1.04 | 1.1 | 1.2 | -1.03 |
| cuticle development | 99 | 1.22 | -1.04 | 1.03 | -1.07 | -1.01 | 1.25 | 1.21 |
| cuticle development involved in protein-based cuticle molting cycle | 13 | 1.28 | -1.16 | -1.04 | -1.21 | -1.09 | 1.14 | 1.17 |
| cuticle development involved in chitin-based cuticle molting cycle | 13 | 1.28 | -1.16 | -1.04 | -1.21 | -1.09 | 1.14 | 1.17 |
| regulation of NF-kappaB import into nucleus | 1 | 1.41 | 1.35 | 1.47 | 1.02 | 1.54 | 2.41 | 1.84 |
| positive regulation of NF-kappaB import into nucleus | 1 | 1.41 | 1.35 | 1.47 | 1.02 | 1.54 | 2.41 | 1.84 |
| GDP-L-fucose biosynthetic process | 2 | -1.67 | 1.21 | 1.13 | 1.24 | 1.36 | -1.89 | -1.14 |
| 'de novo' GDP-L-fucose biosynthetic process | 2 | -1.67 | 1.21 | 1.13 | 1.24 | 1.36 | -1.89 | -1.14 |
| fucose biosynthetic process | 2 | -1.67 | 1.21 | 1.13 | 1.24 | 1.36 | -1.89 | -1.14 |
| L-fucose metabolic process | 2 | -1.67 | 1.21 | 1.13 | 1.24 | 1.36 | -1.89 | -1.14 |
| GDP-4-dehydro-D-rhamnose reductase activity | 1 | 1.27 | 1.21 | 1.12 | 1.11 | 1.24 | -1.28 | 1.03 |
| thiamine diphosphate metabolic process | 1 | 1.06 | 1.62 | -1.26 | 1.22 | -1.14 | -1.11 | 1.12 |
| fat-soluble vitamin biosynthetic process | 2 | 1.28 | 1.03 | 1.33 | -1.0 | 1.21 | 1.44 | 1.16 |
| water-soluble vitamin biosynthetic process | 7 | 1.81 | 1.34 | -1.5 | -1.06 | -1.3 | 1.43 | 1.2 |
| vitamin K biosynthetic process | 1 | 1.46 | 1.06 | 1.55 | -1.07 | 1.35 | 1.82 | 1.43 |
| vitamin K metabolic process | 1 | 1.46 | 1.06 | 1.55 | -1.07 | 1.35 | 1.82 | 1.43 |
| quinone cofactor metabolic process | 6 | 1.17 | 1.16 | 1.19 | 1.16 | 1.13 | 1.18 | -1.0 |
| hemolymph coagulation | 4 | 2.28 | -1.81 | -1.98 | -1.93 | -1.56 | 2.0 | -1.05 |
| sarcolemma | 1 | 1.33 | 1.32 | 2.26 | 1.11 | 1.52 | 1.39 | 1.48 |
| cilium assembly | 33 | -1.14 | 1.06 | 1.0 | 1.05 | -1.01 | -1.08 | -1.05 |
| myosin III complex | 1 | 1.02 | 1.14 | 1.09 | 1.03 | -1.02 | -1.07 | 1.03 |
| hemocyte differentiation | 21 | 1.07 | 1.13 | 1.28 | 1.06 | 1.25 | 1.24 | 1.45 |
| plasmatocyte differentiation | 4 | 1.61 | 1.03 | 1.11 | -1.07 | 1.04 | 1.16 | 1.15 |
| regulation of membrane potential | 25 | 1.35 | 1.01 | -1.06 | -1.33 | -1.11 | 1.68 | 1.19 |
| ecdysis, protein-based cuticle | 7 | 1.02 | 1.08 | 1.04 | -1.01 | -1.03 | 1.01 | 1.06 |
| cellular modified amino acid biosynthetic process | 9 | -1.29 | 1.26 | -1.08 | 1.13 | -1.05 | -1.2 | 1.04 |
| cellular biogenic amine biosynthetic process | 15 | 1.13 | -1.22 | -1.12 | -1.08 | -1.06 | 1.41 | 1.13 |
| cellular biogenic amine catabolic process | 5 | -1.3 | 1.11 | -1.68 | -1.14 | -1.45 | -2.28 | 1.45 |
| dopamine biosynthetic process | 3 | -1.04 | -2.12 | -1.47 | -1.4 | -1.53 | 1.41 | 2.4 |
| dopamine metabolic process | 6 | -1.15 | -1.33 | -1.39 | -1.24 | -1.48 | -1.23 | 1.83 |
| dopamine catabolic process | 1 | -8.98 | 1.77 | -2.05 | -1.02 | -2.08 | -14.22 | 1.22 |
| catecholamine biosynthetic process | 3 | -1.04 | -2.12 | -1.47 | -1.4 | -1.53 | 1.41 | 2.4 |
| catecholamine catabolic process | 1 | -8.98 | 1.77 | -2.05 | -1.02 | -2.08 | -14.22 | 1.22 |
| choline catabolic process | 1 | 2.27 | 1.23 | -5.17 | -1.59 | -2.2 | -9.9 | 9.42 |
| serotonin biosynthetic process | 3 | 1.22 | -1.67 | -1.93 | -1.69 | -1.72 | 1.14 | 2.21 |
| serotonin metabolic process | 4 | -1.49 | -1.27 | -1.96 | -1.49 | -1.8 | -1.76 | 1.9 |
| serotonin catabolic process | 1 | -8.98 | 1.77 | -2.05 | -1.02 | -2.08 | -14.22 | 1.22 |
| indole-containing compound metabolic process | 10 | 1.08 | -1.21 | -1.47 | -1.12 | -1.41 | 1.12 | 1.14 |
| indole-containing compound biosynthetic process | 5 | 1.73 | -1.47 | -1.62 | -1.41 | -1.53 | 1.76 | 1.53 |
| indole-containing compound catabolic process | 3 | -1.86 | 1.07 | -1.41 | -1.17 | -1.45 | -1.76 | -1.05 |
| melanin biosynthetic process | 4 | 1.82 | -1.08 | -1.09 | -1.03 | -1.18 | 1.82 | -1.13 |
| ethanolamine-containing compound metabolic process | 9 | -1.03 | 1.01 | -1.15 | -1.07 | -1.12 | -1.49 | 1.71 |
| pigment metabolic process | 65 | 1.43 | -1.06 | -1.1 | 1.04 | 1.04 | 1.15 | 1.03 |
| eye pigment metabolic process | 30 | 1.05 | -1.06 | -1.17 | 1.17 | 1.04 | -1.09 | -1.16 |
| hormone metabolic process | 36 | 1.14 | -1.24 | -1.31 | -1.23 | -1.11 | -1.09 | 1.2 |
| hormone biosynthetic process | 17 | 1.38 | -1.6 | -1.55 | -1.49 | -1.23 | 1.24 | 1.42 |
| hormone catabolic process | 5 | -1.49 | 1.14 | -1.05 | -1.06 | -1.26 | -2.36 | 1.19 |
| arginine biosynthetic process via ornithine | 1 | -15.39 | 1.51 | -7.96 | 1.39 | 1.15 | -4.77 | -1.11 |
| purine nucleoside biosynthetic process | 7 | 1.07 | 1.09 | -1.03 | 1.13 | 1.19 | -1.05 | -1.12 |
| ribonucleoside catabolic process | 3 | -2.8 | 1.04 | -1.39 | 1.2 | 1.32 | -1.87 | -1.06 |
| ribonucleoside biosynthetic process | 7 | 1.07 | 1.09 | -1.03 | 1.13 | 1.19 | -1.05 | -1.12 |
| photoreceptor cell development | 72 | 1.13 | 1.13 | 1.31 | 1.04 | 1.26 | 1.19 | 1.22 |
| eye photoreceptor cell development | 61 | 1.11 | 1.13 | 1.34 | 1.04 | 1.25 | 1.22 | 1.23 |
| ocellus photoreceptor cell development | 2 | -1.04 | 1.07 | -1.15 | -1.03 | -1.15 | -1.11 | 1.0 |
| regulation of eye photoreceptor cell development | 11 | 1.05 | 1.08 | 1.34 | 1.01 | 1.28 | 1.07 | 1.22 |
| positive regulation of eye photoreceptor cell development | 1 | 2.02 | 1.25 | 1.56 | 1.13 | 1.49 | 2.5 | 1.69 |
| negative regulation of eye photoreceptor cell development | 3 | -1.04 | -1.04 | 1.09 | -1.03 | 1.27 | -1.29 | 1.13 |
| aspartic endopeptidase activity, intramembrane cleaving | 2 | -1.09 | 1.35 | -1.07 | 1.16 | -1.03 | -1.2 | 1.46 |
| regulation of tyrosine phosphorylation of STAT protein | 1 | 1.28 | 1.03 | 1.08 | -1.21 | -1.11 | 1.23 | -1.01 |
| negative regulation of tyrosine phosphorylation of STAT protein | 1 | 1.28 | 1.03 | 1.08 | -1.21 | -1.11 | 1.23 | -1.01 |
| benzene-containing compound metabolic process | 9 | -1.02 | -1.18 | -1.26 | -1.16 | -1.28 | -1.1 | 1.52 |
| hyperosmotic salinity response | 1 | 1.22 | 1.33 | 2.18 | 1.07 | 1.72 | 2.05 | 1.61 |
| response to hydrogen peroxide | 9 | -1.09 | 1.06 | -1.06 | 1.22 | 1.22 | -1.32 | -1.19 |
| neuron maturation | 19 | 1.17 | -1.01 | 1.21 | 1.01 | 1.4 | 1.66 | 1.06 |
| pteridine-containing compound metabolic process | 19 | 1.66 | 1.0 | 1.01 | 1.02 | -1.03 | 1.17 | 1.15 |
| pteridine-containing compound biosynthetic process | 17 | 1.71 | -1.0 | -1.01 | -1.01 | -1.05 | 1.15 | 1.18 |
| pteridine-containing compound catabolic process | 1 | -1.35 | 1.22 | 1.7 | 1.18 | 1.15 | -1.09 | 1.03 |
| hormone binding | 9 | 1.25 | 1.01 | 1.03 | 1.03 | 1.09 | 1.15 | 1.08 |
| retinol metabolic process | 1 | -1.07 | 1.02 | 1.03 | -1.03 | -1.28 | 1.11 | -1.06 |
| retinal metabolic process | 2 | 1.02 | 1.01 | 1.08 | 1.02 | -1.08 | 1.12 | -1.06 |
| DNA polymerase complex | 7 | 1.2 | 1.16 | 1.12 | 1.11 | 1.16 | 1.17 | -1.15 |
| lipid phosphatase activity | 2 | -1.51 | 1.0 | 1.16 | 1.09 | 1.56 | -1.48 | -1.22 |
| phosphoric ester hydrolase activity | 206 | 1.06 | 1.0 | -1.06 | 1.02 | 1.1 | 1.08 | 1.0 |
| microbody | 39 | 1.3 | -1.06 | -1.53 | 1.05 | 1.16 | -1.5 | -1.16 |
| peptide deformylase activity | 2 | 1.03 | 1.02 | -2.05 | 1.3 | 1.27 | -1.09 | -2.35 |
| homeostatic process | 183 | 1.07 | 1.02 | 1.12 | -1.0 | 1.08 | 1.18 | 1.06 |
| glucose homeostasis | 1 | 1.23 | 1.27 | 1.15 | 1.19 | 1.2 | 1.15 | 1.07 |
| response to starvation | 29 | 1.1 | -1.06 | 1.01 | -1.06 | 1.08 | -1.01 | 1.16 |
| behavioral response to starvation | 4 | -1.14 | -2.62 | -2.82 | -2.72 | -3.01 | -3.18 | 1.82 |
| periplasmic space | 9 | 1.87 | -1.07 | 1.01 | -1.1 | -1.04 | 1.24 | -1.01 |
| vesicular fraction | 85 | -1.05 | -1.16 | -1.3 | -1.04 | 1.03 | -1.24 | -1.12 |
| chorion | 17 | 1.11 | -1.0 | -1.02 | 1.07 | -1.04 | -1.16 | -1.04 |
| ATPase activity, coupled to transmembrane movement of ions | 67 | -1.16 | 1.01 | 1.26 | -1.1 | -1.0 | 1.35 | -1.07 |
| mating plug formation | 2 | 1.04 | 1.05 | 1.11 | 1.02 | -1.07 | -1.15 | -1.16 |
| cholesterol homeostasis | 3 | -6.81 | -2.38 | -1.29 | 1.48 | -2.64 | -3.47 | -5.64 |
| actomyosin | 1 | 1.76 | 1.07 | 1.13 | 1.08 | 1.49 | 1.28 | 1.15 |
| mitochondrial nucleoid | 1 | 1.18 | 1.16 | 1.75 | 1.38 | 1.68 | 1.06 | -1.24 |
| ecdysis-triggering hormone receptor activity | 1 | -1.33 | -1.06 | -1.14 | -1.28 | -1.36 | -1.06 | -1.24 |
| JUN kinase kinase kinase kinase activity | 1 | -1.25 | 1.21 | 1.4 | 1.19 | 1.07 | 1.11 | 1.44 |
| regulation of cell fate specification | 25 | 1.18 | 1.07 | 1.15 | 1.01 | 1.2 | 1.06 | 1.15 |
| regulation of retinal cone cell fate specification | 2 | -1.43 | 1.42 | 1.17 | 1.08 | 1.59 | -1.55 | 1.31 |
| compound eye cone cell differentiation | 14 | 1.25 | 1.17 | 1.35 | 1.14 | 1.48 | 1.22 | 1.4 |
| compound eye cone cell fate commitment | 7 | 1.02 | 1.24 | 1.18 | 1.18 | 1.59 | 1.22 | 1.33 |
| regulation of compound eye cone cell fate specification | 2 | 1.2 | 1.07 | 1.13 | -1.09 | 1.23 | 1.23 | 1.3 |
| negative regulation of compound eye cone cell fate specification | 2 | 1.2 | 1.07 | 1.13 | -1.09 | 1.23 | 1.23 | 1.3 |
| cardioblast cell fate commitment | 6 | 1.11 | 1.0 | 1.08 | 1.01 | 1.1 | -1.11 | 1.03 |
| cardioblast cell fate specification | 1 | -1.42 | 1.01 | 1.2 | 1.29 | 2.44 | -1.59 | 1.35 |
| regulation of cardioblast cell fate specification | 8 | 1.02 | 1.09 | 1.19 | 1.04 | 1.21 | -1.01 | 1.39 |
| crystal cell differentiation | 8 | 1.09 | 1.04 | 1.08 | 1.05 | 1.19 | 1.34 | 1.82 |
| regulation of crystal cell differentiation | 7 | 1.04 | 1.03 | 1.09 | 1.03 | 1.17 | 1.42 | 1.89 |
| negative regulation of crystal cell differentiation | 4 | 1.02 | 1.05 | 1.07 | 1.04 | 1.02 | 1.08 | 1.37 |
| positive regulation of crystal cell differentiation | 1 | -1.18 | -1.04 | -1.07 | -1.26 | 1.04 | 1.06 | 5.07 |
| muscle cell differentiation | 115 | 1.29 | 1.03 | 1.23 | -1.03 | 1.17 | 1.35 | 1.21 |
| muscle cell fate commitment | 8 | -1.1 | -1.16 | -1.07 | -1.18 | -1.07 | -1.08 | 2.39 |
| muscle cell fate specification | 3 | 1.42 | 1.19 | 1.29 | 1.07 | 1.29 | 2.0 | 1.68 |
| ovulation cycle | 1 | -1.49 | 2.58 | -1.26 | -1.23 | -1.23 | -1.38 | 1.24 |
| ocellus photoreceptor cell differentiation | 2 | -1.04 | 1.07 | -1.15 | -1.03 | -1.15 | -1.11 | 1.0 |
| eye photoreceptor cell fate commitment | 51 | 1.32 | 1.09 | 1.23 | 1.09 | 1.32 | 1.24 | 1.18 |
| succinate-CoA ligase complex | 4 | 1.26 | -1.07 | 1.22 | -1.12 | 1.17 | 1.73 | -1.33 |
| dosage compensation complex assembly | 3 | 1.47 | 1.24 | 1.5 | 1.08 | 1.32 | 1.62 | 1.47 |
| mitochondrial intermembrane space protein transporter complex | 6 | 1.1 | 1.08 | 1.31 | 1.16 | 1.2 | 1.34 | -1.18 |
| thiamine-containing compound metabolic process | 1 | 1.06 | 1.62 | -1.26 | 1.22 | -1.14 | -1.11 | 1.12 |
| thiamine-containing compound biosynthetic process | 1 | 1.06 | 1.62 | -1.26 | 1.22 | -1.14 | -1.11 | 1.12 |
| flavin-containing compound metabolic process | 2 | 1.78 | 2.05 | -2.11 | -2.16 | -2.52 | 2.14 | 1.44 |
| flavin-containing compound biosynthetic process | 2 | 1.78 | 2.05 | -2.11 | -2.16 | -2.52 | 2.14 | 1.44 |
| D-xylose metabolic process | 1 | 1.17 | 1.3 | 1.39 | 1.35 | 1.31 | 1.22 | 1.24 |
| hydrogen peroxide metabolic process | 5 | -1.31 | 1.04 | -1.13 | 1.14 | 1.05 | -1.39 | -1.21 |
| hydrogen peroxide catabolic process | 5 | -1.31 | 1.04 | -1.13 | 1.14 | 1.05 | -1.39 | -1.21 |
| circadian sleep/wake cycle | 19 | -1.16 | 1.13 | 1.1 | -1.04 | 1.16 | 1.12 | 1.12 |
| regulation of circadian sleep/wake cycle | 16 | -1.12 | 1.18 | 1.14 | -1.02 | 1.04 | 1.13 | 1.15 |
| regulation of circadian rhythm | 34 | 1.14 | 1.12 | 1.18 | 1.01 | 1.11 | 1.4 | 1.23 |
| positive regulation of circadian rhythm | 2 | 1.24 | 1.16 | 1.04 | -1.02 | 1.05 | 1.25 | 1.29 |
| drinking behavior | 2 | -1.36 | -1.02 | 1.18 | 1.1 | 1.66 | -1.79 | 1.09 |
| long-chain fatty acid biosynthetic process | 1 | 12.72 | -1.88 | -1.16 | -3.3 | -1.54 | 3.13 | 2.87 |
| very long-chain fatty acid biosynthetic process | 1 | 12.72 | -1.88 | -1.16 | -3.3 | -1.54 | 3.13 | 2.87 |
| GPI-anchor transamidase complex | 2 | 1.49 | 1.08 | 1.21 | 1.27 | 1.12 | -1.05 | -1.02 |
| nucleosome mobilization | 5 | 1.33 | 1.2 | 1.59 | 1.27 | 1.63 | 1.57 | 1.26 |
| ecdysteroid 22-hydroxylase activity | 1 | -1.29 | -1.12 | -1.07 | -1.16 | -1.36 | -1.02 | -1.2 |
| ecdysteroid 2-hydroxylase activity | 1 | -1.19 | 1.02 | -1.06 | 1.03 | -1.14 | -1.03 | -1.16 |
| signal transduction in response to DNA damage | 9 | 1.53 | 1.18 | 1.1 | 1.13 | 1.27 | 1.31 | 1.44 |
| DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis | 2 | 1.7 | 1.37 | 1.03 | 1.24 | 1.62 | 1.04 | 1.2 |
| ATP synthesis coupled electron transport | 59 | 1.05 | 1.03 | 1.22 | 1.05 | 1.09 | 1.56 | -1.33 |
| mitochondrial ATP synthesis coupled electron transport | 57 | 1.06 | 1.03 | 1.23 | 1.05 | 1.1 | 1.56 | -1.33 |
| tRNA 3'-trailer cleavage | 1 | 1.37 | 1.18 | 1.82 | 1.25 | 1.62 | 1.5 | 1.42 |
| tRNA 3'-end processing | 1 | 1.37 | 1.18 | 1.82 | 1.25 | 1.62 | 1.5 | 1.42 |
| snRNA transcription from RNA polymerase II promoter | 4 | 1.14 | 1.21 | 1.28 | 1.12 | 1.26 | 1.23 | 1.22 |
| snRNA transcription from RNA polymerase III promoter | 3 | 1.69 | 1.17 | 1.43 | 1.09 | 1.41 | 1.27 | 1.23 |
| tRNA transcription from RNA polymerase III promoter | 1 | 1.67 | 1.25 | 1.56 | 1.07 | 1.53 | 1.47 | 1.26 |
| histone methyltransferase activity (H4-K20 specific) | 3 | 1.11 | 1.29 | 1.61 | 1.13 | 1.69 | 1.86 | 1.45 |
| histone methyltransferase activity (H3-K4 specific) | 4 | 1.72 | 1.25 | 1.86 | 1.03 | 1.8 | 2.11 | 1.88 |
| polo kinase kinase activity | 1 | 1.11 | 1.2 | 3.72 | 1.08 | 1.65 | 2.06 | 2.0 |
| protein homodimerization activity | 65 | 1.35 | 1.04 | 1.0 | -1.02 | 1.11 | 1.22 | 1.18 |
| protein homooligomerization | 4 | 1.26 | 1.29 | 1.33 | 1.14 | 1.48 | 1.55 | 1.11 |
| actinin binding | 1 | 1.14 | -1.01 | 1.12 | 1.41 | 1.03 | -1.2 | -1.43 |
| fucose binding | 2 | 1.18 | -1.09 | -1.2 | -1.39 | -1.4 | -1.1 | 2.98 |
| pheromone metabolic process | 9 | 1.62 | -1.49 | -1.71 | -1.43 | -1.39 | 1.17 | 1.34 |
| pheromone biosynthetic process | 3 | 1.42 | -3.74 | -5.02 | -3.4 | -2.87 | -1.24 | 2.33 |
| vitamin B6 metabolic process | 4 | 2.09 | 1.04 | -1.32 | 1.26 | 1.04 | 1.32 | 1.12 |
| vitamin B6 biosynthetic process | 4 | 2.09 | 1.04 | -1.32 | 1.26 | 1.04 | 1.32 | 1.12 |
| histone deacetylase binding | 13 | 2.66 | -1.15 | 1.12 | 1.01 | 1.08 | 1.67 | 1.03 |
| peptidoglycan binding | 12 | -1.13 | -1.64 | -1.19 | -1.04 | -1.82 | -1.69 | -1.29 |
| amide transport | 1 | 1.41 | 1.17 | -1.02 | 1.18 | -1.05 | -1.09 | 1.18 |
| xenobiotic transporter activity | 8 | -1.32 | 1.18 | 1.1 | 1.25 | 1.08 | -1.09 | 1.15 |
| D-amino acid transport | 1 | -1.04 | -22.78 | -24.12 | -1.99 | 7.91 | 4.55 | 1.12 |
| D-amino acid transmembrane transporter activity | 1 | -1.04 | -22.78 | -24.12 | -1.99 | 7.91 | 4.55 | 1.12 |
| glucoside transport | 2 | 1.24 | -2.16 | -1.19 | -2.0 | -1.13 | 3.38 | 1.3 |
| lipoprotein transport | 1 | -1.18 | 2.03 | -1.34 | -1.0 | -1.31 | -1.12 | -1.14 |
| ornithine decarboxylase regulator activity | 1 | 1.05 | 1.03 | -1.12 | 1.06 | -1.03 | -1.01 | 1.05 |
| amyloid precursor protein metabolic process | 1 | 1.02 | 1.3 | 1.25 | 1.11 | 1.03 | 1.74 | 1.33 |
| regulation of transcription factor import into nucleus | 22 | 1.33 | 1.06 | 1.24 | 1.08 | 1.34 | 1.31 | 1.23 |
| negative regulation of transcription factor import into nucleus | 4 | 2.74 | 1.08 | 1.33 | 1.02 | 1.2 | 2.19 | 1.75 |
| positive regulation of transcription factor import into nucleus | 18 | 1.13 | 1.06 | 1.22 | 1.1 | 1.37 | 1.17 | 1.13 |
| cell projection | 159 | 1.04 | 1.05 | 1.18 | -1.0 | 1.11 | 1.26 | 1.13 |
| neuron projection | 92 | 1.14 | 1.04 | 1.23 | -1.01 | 1.17 | 1.39 | 1.21 |
| maintenance of rDNA | 1 | 1.01 | 1.1 | 1.09 | 1.14 | -1.01 | 1.17 | 1.11 |
| camera-type eye development | 1 | -1.1 | -1.15 | -1.07 | -1.18 | 1.52 | 1.28 | 1.24 |
| gamma-tubulin binding | 5 | 1.28 | 1.03 | 1.2 | 1.09 | 1.23 | 1.33 | 1.14 |
| ribonucleoprotein complex binding | 11 | 1.27 | 1.17 | 1.32 | 1.05 | 1.14 | 1.31 | 1.33 |
| ribosome binding | 6 | 1.66 | 1.06 | 1.42 | 1.03 | 1.28 | 1.53 | 1.23 |
| ribosomal small subunit binding | 1 | 2.01 | 1.25 | 1.56 | -1.2 | 1.13 | 3.31 | 1.74 |
| neuronal cell body | 26 | -1.0 | -1.02 | 1.2 | 1.05 | 1.13 | 1.07 | 1.02 |
| cysteine-type endopeptidase inhibitor activity involved in apoptotic process | 1 | -1.96 | -1.03 | 2.81 | 1.35 | 1.53 | -2.84 | -1.09 |
| cysteine-type endopeptidase regulator activity involved in apoptotic process | 1 | -1.96 | -1.03 | 2.81 | 1.35 | 1.53 | -2.84 | -1.09 |
| costamere | 1 | 1.22 | 1.1 | -1.01 | 1.09 | -1.06 | -1.14 | -1.04 |
| amino acid activation | 42 | -1.06 | 1.25 | 1.26 | 1.14 | 1.18 | 1.05 | 1.08 |
| tRNA aminoacylation | 41 | -1.07 | 1.25 | 1.27 | 1.14 | 1.19 | 1.05 | 1.09 |
| amino acid activation for nonribosomal peptide biosynthetic process | 1 | 1.36 | 1.09 | 1.02 | -1.03 | -1.16 | 1.04 | 1.01 |
| amino acid adenylylation by nonribosomal peptide synthase | 1 | 1.36 | 1.09 | 1.02 | -1.03 | -1.16 | 1.04 | 1.01 |
| peptide biosynthetic process | 4 | -2.1 | 1.35 | -1.25 | 1.15 | -1.28 | -1.79 | 1.11 |
| ATP-dependent chromatin remodeling | 11 | 1.15 | 1.19 | 1.36 | 1.14 | 1.3 | 1.34 | 1.2 |
| DNA methylation involved in embryo development | 1 | 1.54 | 1.07 | -1.03 | 1.11 | 1.17 | 1.09 | 1.04 |
| thermotaxis | 16 | 1.05 | -1.29 | -1.09 | -1.04 | 1.16 | -1.11 | -1.04 |
| regulation of forward locomotion | 1 | 1.3 | 1.0 | -1.05 | -1.09 | 1.04 | -1.06 | -1.0 |
| meiotic metaphase I plate congression | 1 | -1.17 | 1.02 | -1.13 | 1.11 | -1.02 | -1.2 | -1.13 |
| extracellular structure organization | 38 | -1.02 | 1.05 | 1.2 | -1.0 | 1.18 | 1.05 | 1.23 |
| intercellular bridge organization | 13 | 1.05 | 1.09 | 1.3 | 1.08 | 1.37 | 1.26 | 1.26 |
| flagellum organization | 10 | -1.03 | 1.09 | 1.07 | 1.09 | 1.11 | 1.01 | -1.02 |
| positive regulation of apoptotic process | 34 | 1.1 | 1.17 | 1.11 | 1.1 | 1.27 | 1.06 | 1.22 |
| negative regulation of apoptotic process | 49 | 1.26 | 1.09 | 1.28 | 1.03 | 1.27 | 1.36 | 1.28 |
| regulation of programmed cell death | 113 | 1.08 | 1.13 | 1.31 | 1.07 | 1.3 | 1.2 | 1.21 |
| positive regulation of programmed cell death | 49 | -1.01 | 1.15 | 1.2 | 1.09 | 1.32 | 1.04 | 1.24 |
| negative regulation of programmed cell death | 55 | 1.22 | 1.09 | 1.3 | 1.02 | 1.24 | 1.34 | 1.26 |
| germ cell nucleus | 10 | -1.1 | 1.13 | 1.01 | 1.03 | -1.05 | -1.24 | 1.05 |
| synaptic cleft | 1 | 3.21 | -1.32 | 1.06 | -1.26 | 1.51 | 5.17 | 1.89 |
| positive regulation of catalytic activity | 62 | 1.12 | 1.13 | 1.26 | 1.11 | 1.27 | 1.29 | 1.11 |
| negative regulation of catalytic activity | 22 | 1.29 | 1.1 | 1.29 | 1.14 | 1.27 | 1.24 | 1.11 |
| regulation of GTPase activity | 42 | 1.08 | 1.16 | 1.35 | 1.05 | 1.29 | 1.31 | 1.21 |
| amino acid import | 4 | -2.33 | 1.09 | -2.25 | -1.01 | 1.43 | -1.47 | -1.72 |
| L-amino acid import | 1 | -51.3 | 1.56 | -15.93 | -1.24 | 2.78 | -3.27 | -134.18 |
| cellular metabolic compound salvage | 6 | 1.62 | 1.11 | -1.15 | 1.11 | 1.11 | 1.24 | -1.06 |
| regulation of GTP cyclohydrolase I activity | 2 | 6.83 | -2.51 | -2.16 | -2.15 | -1.79 | -1.27 | 2.69 |
| purine base salvage | 1 | 1.17 | 1.37 | 1.23 | 1.24 | 1.3 | 1.33 | -1.5 |
| purine-containing compound salvage | 4 | 1.03 | 1.11 | -1.11 | 1.23 | 1.2 | 1.03 | -1.31 |
| amino acid salvage | 1 | 1.92 | 1.16 | 1.15 | 1.28 | 1.06 | 2.87 | -1.21 |
| positive regulation of GTP cyclohydrolase I activity | 1 | 1.39 | -1.12 | 1.24 | 1.31 | 1.42 | -1.23 | 1.04 |
| receptor metabolic process | 7 | 1.34 | -1.03 | 1.06 | -1.07 | 1.17 | 1.5 | 1.02 |
| regulation of I-kappaB kinase/NF-kappaB cascade | 2 | 1.37 | 1.19 | 1.03 | 1.23 | 1.17 | 1.39 | -1.06 |
| negative regulation of I-kappaB kinase/NF-kappaB cascade | 1 | 1.33 | 1.06 | 1.15 | 1.25 | 1.2 | 1.46 | -1.1 |
| ubiquitin binding | 2 | 1.76 | 1.35 | 1.96 | 1.2 | 1.97 | 1.51 | 1.56 |
| 3'-5' DNA helicase activity | 12 | -1.05 | 1.14 | 1.22 | 1.27 | 1.49 | -1.11 | -1.06 |
| ATP-dependent 3'-5' DNA helicase activity | 2 | -1.1 | 1.25 | 1.24 | 1.2 | 1.14 | 1.04 | 1.15 |
| single-stranded DNA-dependent ATPase activity | 3 | 1.08 | 1.14 | 1.41 | 1.15 | 1.37 | -1.0 | 1.15 |
| spindle stabilization | 9 | 1.1 | 1.19 | 1.29 | 1.13 | 1.34 | 1.33 | 1.14 |
| meiotic spindle stabilization | 4 | 1.18 | 1.25 | 1.5 | 1.11 | 1.42 | 1.76 | 1.44 |
| mitotic spindle stabilization | 4 | 1.04 | 1.15 | 1.21 | 1.11 | 1.33 | 1.1 | -1.02 |
| induction of bacterial agglutination | 1 | -1.2 | -1.21 | 1.01 | 1.71 | 2.04 | -1.29 | -1.16 |
| entrainment of circadian clock by photoperiod | 6 | 1.21 | 1.05 | -1.08 | 1.13 | -1.05 | 1.32 | 1.05 |
| negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 1.22 | -1.04 | 1.69 | -1.06 | 1.27 | -1.09 | -1.19 |
| proteasomal ubiquitin-dependent protein catabolic process | 17 | 1.04 | 1.17 | 1.09 | 1.09 | 1.14 | -1.02 | 1.15 |
| ion binding | 1429 | -1.0 | 1.0 | 1.0 | 1.02 | 1.08 | 1.03 | 1.02 |
| anion binding | 1 | 1.12 | 1.0 | 1.13 | 1.06 | 1.1 | -1.01 | -1.1 |
| cation binding | 1428 | -1.0 | 1.0 | 1.0 | 1.02 | 1.08 | 1.03 | 1.02 |
| macromolecule metabolic process | 2469 | -1.04 | -1.01 | 1.03 | 1.06 | 1.1 | -1.03 | -1.03 |
| nucleotide salvage | 1 | 8.31 | 1.05 | -1.79 | -1.54 | -1.19 | 1.17 | 2.54 |
| nucleoside salvage | 4 | 1.03 | 1.11 | -1.11 | 1.23 | 1.2 | 1.03 | -1.31 |
| amine binding | 16 | -1.41 | -1.08 | -1.46 | 1.15 | 1.12 | -1.1 | -1.47 |
| alcohol binding | 1 | 1.12 | 1.0 | 1.13 | 1.06 | 1.1 | -1.01 | -1.1 |
| rhythmic excitation | 1 | 9.13 | 1.03 | 1.44 | -1.07 | -1.11 | 1.07 | 1.06 |
| P granule | 13 | 1.42 | 1.03 | 1.1 | 1.02 | 1.09 | 1.51 | -1.01 |
| H4/H2A histone acetyltransferase complex | 6 | 1.43 | 1.08 | 1.23 | 1.19 | 1.24 | 1.41 | -1.02 |
| terminal button | 22 | 1.26 | 1.08 | 1.07 | -1.18 | 1.13 | 1.67 | 1.31 |
| varicosity | 1 | -1.06 | -1.47 | -1.01 | -1.23 | -1.12 | 1.54 | -1.14 |
| dendritic spine | 4 | 1.47 | 1.02 | 1.26 | -1.09 | 1.32 | 1.72 | 1.43 |
| response to amino acid stimulus | 4 | 1.59 | -1.01 | 1.38 | 1.29 | 1.34 | 1.2 | -1.18 |
| perikaryon | 1 | -1.51 | 1.58 | 1.98 | 1.08 | 1.74 | 1.62 | 1.22 |
| anion transmembrane-transporting ATPase activity | 1 | 1.35 | 1.29 | 1.36 | 1.08 | 1.19 | 1.34 | 1.11 |
| organelle | 3099 | 1.13 | 1.07 | 1.15 | 1.08 | 1.18 | 1.16 | 1.04 |
| membrane-bounded organelle | 2551 | 1.12 | 1.08 | 1.16 | 1.09 | 1.18 | 1.16 | 1.04 |
| non-membrane-bounded organelle | 1053 | 1.16 | 1.07 | 1.18 | 1.08 | 1.22 | 1.19 | 1.04 |
| intracellular organelle | 3091 | 1.13 | 1.07 | 1.15 | 1.08 | 1.18 | 1.16 | 1.04 |
| intracellular membrane-bounded organelle | 2548 | 1.12 | 1.08 | 1.16 | 1.09 | 1.18 | 1.16 | 1.03 |
| intracellular non-membrane-bounded organelle | 1053 | 1.16 | 1.07 | 1.18 | 1.08 | 1.22 | 1.19 | 1.04 |
| organelle lumen | 588 | 1.2 | 1.12 | 1.23 | 1.16 | 1.24 | 1.22 | -1.0 |
| protein complex | 1479 | 1.14 | 1.07 | 1.19 | 1.07 | 1.18 | 1.18 | 1.05 |
| receptor complex | 33 | -1.32 | 1.13 | 1.15 | 1.02 | 1.11 | -1.17 | 1.16 |
| protein complex disassembly | 15 | -1.02 | 1.15 | 1.27 | 1.13 | 1.29 | 1.16 | 1.02 |
| negative regulation of protein complex disassembly | 17 | 1.02 | 1.13 | 1.55 | 1.04 | 1.43 | 1.45 | 1.22 |
| positive regulation of protein complex disassembly | 2 | 1.03 | 1.18 | 1.17 | -1.04 | 1.37 | 1.47 | 1.3 |
| regulation of protein complex disassembly | 20 | 1.04 | 1.15 | 1.45 | 1.04 | 1.4 | 1.38 | 1.24 |
| proteasome assembly | 2 | 1.37 | 1.16 | 1.24 | 1.14 | 1.1 | -1.0 | 1.34 |
| regulation of protein complex assembly | 23 | -1.13 | 1.08 | 1.36 | 1.07 | 1.31 | -1.05 | 1.24 |
| laminin complex | 1 | 3.52 | 1.3 | 1.81 | 1.23 | 2.46 | 6.46 | 1.6 |
| regulation of ion transport | 23 | 1.04 | 1.04 | 1.16 | 1.01 | 1.04 | 1.18 | 1.01 |
| positive regulation of ion transport | 7 | 1.34 | 1.0 | 1.16 | 1.0 | 1.09 | 1.43 | 1.16 |
| negative regulation of ion transport | 1 | -1.32 | 1.16 | 1.21 | -1.14 | -1.21 | 1.47 | 1.08 |
| apoptotic cell clearance | 8 | 1.13 | -1.17 | 1.05 | -1.1 | 1.15 | 1.39 | 1.06 |
| response to alkaloid | 20 | 1.2 | 1.11 | 1.03 | -1.07 | 1.16 | 1.49 | 1.28 |
| positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 14 | 1.01 | 1.14 | 1.25 | 1.19 | 1.42 | 1.19 | 1.06 |
| regulation of cysteine-type endopeptidase activity involved in apoptotic process | 18 | 1.12 | 1.11 | 1.23 | 1.18 | 1.35 | 1.14 | 1.01 |
| apocarotenoid metabolic process | 2 | 1.02 | 1.01 | 1.08 | 1.02 | -1.08 | 1.12 | -1.06 |
| glutathione binding | 1 | 5.44 | 1.27 | 1.83 | 1.24 | 1.81 | 2.29 | 1.39 |
| apical junction complex | 45 | 1.96 | -1.09 | 1.26 | -1.14 | 1.29 | 2.5 | 1.48 |
| apical junction assembly | 33 | 2.06 | -1.11 | 1.37 | -1.16 | 1.26 | 2.36 | 1.49 |
| pigment metabolic process involved in developmental pigmentation | 30 | 1.05 | -1.06 | -1.17 | 1.17 | 1.04 | -1.09 | -1.16 |
| response to dsRNA | 11 | 1.46 | 1.17 | 1.45 | 1.09 | 1.32 | 1.46 | 1.31 |
| positive regulation of DNA binding | 1 | 2.81 | 1.45 | 1.89 | 1.09 | 1.48 | 2.58 | 1.71 |
| negative regulation of DNA binding | 3 | 1.1 | 1.28 | 1.83 | 1.03 | 1.28 | 1.14 | 1.67 |
| regulation of protein binding | 1 | 1.3 | 1.31 | 1.72 | 1.5 | 1.61 | 1.9 | 1.2 |
| proteoglycan binding | 1 | 3.21 | -1.21 | 1.67 | 1.12 | 1.32 | 6.4 | 2.23 |
| heparan sulfate proteoglycan binding | 1 | 3.21 | -1.21 | 1.67 | 1.12 | 1.32 | 6.4 | 2.23 |
| steroid hormone mediated signaling pathway | 19 | 1.11 | 1.08 | 1.02 | -1.0 | 1.09 | 1.16 | 1.16 |
| regulation of MAP kinase activity | 15 | 1.26 | 1.16 | 1.47 | 1.13 | 1.4 | 1.82 | 1.2 |
| positive regulation of MAP kinase activity | 9 | 1.14 | 1.19 | 1.51 | 1.11 | 1.38 | 1.84 | 1.15 |
| negative regulation of MAP kinase activity | 6 | 1.46 | 1.13 | 1.41 | 1.17 | 1.43 | 1.78 | 1.28 |
| regulation of MAPK cascade | 39 | 1.06 | 1.19 | 1.25 | 1.1 | 1.26 | 1.33 | 1.23 |
| negative regulation of MAPK cascade | 14 | 1.21 | 1.15 | 1.44 | 1.11 | 1.27 | 1.31 | 1.15 |
| positive regulation of MAPK cascade | 15 | 1.07 | 1.22 | 1.31 | 1.02 | 1.16 | 1.33 | 1.35 |
| macromolecule modification | 665 | 1.18 | 1.1 | 1.22 | 1.08 | 1.22 | 1.25 | 1.13 |
| macromolecule glycosylation | 51 | 1.14 | 1.04 | 1.06 | 1.04 | 1.1 | 1.12 | 1.22 |
| macromolecule methylation | 44 | 1.36 | 1.18 | 1.35 | 1.17 | 1.37 | 1.34 | 1.16 |
| bHLH transcription factor binding | 1 | 1.46 | 1.06 | -1.06 | 1.12 | -1.04 | -1.02 | 1.13 |
| negative regulation of sequence-specific DNA binding transcription factor activity | 4 | 2.01 | 1.08 | 1.16 | 1.02 | 1.02 | 1.35 | 1.88 |
| response to peptide hormone stimulus | 20 | -1.09 | 1.13 | 1.2 | 1.13 | 1.18 | 1.07 | 1.2 |
| oxoacid metabolic process | 239 | -1.01 | -1.01 | -1.15 | -1.03 | 1.0 | 1.06 | -1.05 |
| cellular alkane metabolic process | 1 | 1.08 | 1.05 | 1.26 | 1.07 | 1.31 | 1.33 | 1.1 |
| alkane biosynthetic process | 1 | 1.08 | 1.05 | 1.26 | 1.07 | 1.31 | 1.33 | 1.1 |
| cellular alkene metabolic process | 1 | -1.74 | 1.11 | -2.01 | 1.51 | 1.27 | -1.87 | -1.55 |
| alkene biosynthetic process | 1 | -1.74 | 1.11 | -2.01 | 1.51 | 1.27 | -1.87 | -1.55 |
| regulation of secondary metabolic process | 6 | 1.65 | -1.7 | -1.31 | -1.28 | -1.22 | 1.14 | -1.32 |
| proton-transporting ATP synthase complex assembly | 2 | 1.21 | 1.11 | 1.39 | 1.2 | 1.21 | 1.72 | -1.33 |
| regulation of ATPase activity | 1 | -1.06 | 1.13 | 1.64 | 1.31 | 1.74 | 1.65 | 1.23 |
| regulation of generation of precursor metabolites and energy | 1 | -2.86 | 1.25 | -1.2 | 1.45 | 1.26 | 1.29 | -1.32 |
| pigmentation | 60 | 1.18 | -1.08 | -1.08 | 1.07 | 1.0 | -1.05 | 1.0 |
| pigment metabolic process involved in pigmentation | 30 | 1.05 | -1.06 | -1.17 | 1.17 | 1.04 | -1.09 | -1.16 |
| regulation of RNA splicing | 60 | 1.18 | 1.18 | 1.27 | 1.17 | 1.24 | 1.38 | 1.08 |
| regulation of RNA stability | 2 | 2.6 | 1.13 | 1.81 | 1.28 | 1.81 | 2.02 | 1.35 |
| regulation of mRNA stability | 2 | 2.6 | 1.13 | 1.81 | 1.28 | 1.81 | 2.02 | 1.35 |
| ATPase activity, coupled to movement of substances | 120 | -1.08 | -1.04 | 1.03 | -1.07 | 1.02 | 1.16 | -1.05 |
| cell surface binding | 17 | 1.52 | -1.27 | -1.19 | -1.21 | -1.12 | 1.67 | 1.0 |
| eukaryotic cell surface binding | 2 | 2.1 | -1.18 | 1.11 | -1.38 | 1.18 | 2.66 | 3.0 |
| regulation of JUN kinase activity | 7 | 1.24 | 1.11 | 1.42 | 1.16 | 1.39 | 1.42 | 1.27 |
| positive regulation of JUN kinase activity | 3 | -1.04 | 1.28 | 1.57 | 1.15 | 1.45 | 1.3 | 1.45 |
| negative regulation of JUN kinase activity | 4 | 1.5 | -1.01 | 1.32 | 1.18 | 1.35 | 1.51 | 1.15 |
| regulation of neuron apoptotic process | 11 | 1.33 | 1.08 | 1.15 | -1.03 | 1.14 | 1.6 | 1.39 |
| negative regulation of neuron apoptotic process | 11 | 1.33 | 1.08 | 1.15 | -1.03 | 1.14 | 1.6 | 1.39 |
| protein serine/threonine kinase activator activity | 2 | 1.16 | 1.13 | -1.02 | 1.22 | 1.1 | -1.36 | -1.19 |
| protein acylation | 39 | 1.04 | 1.16 | 1.27 | 1.15 | 1.2 | 1.05 | 1.04 |
| molybdopterin cofactor metabolic process | 5 | 1.14 | 1.31 | 1.37 | 1.14 | 1.16 | -1.34 | 1.2 |
| molybdopterin cofactor binding | 1 | 1.38 | 1.39 | -1.09 | 1.59 | 1.39 | -1.89 | 1.34 |
| positive regulation of GTPase activity | 5 | 1.0 | 1.24 | 1.63 | 1.21 | 1.82 | 1.58 | 1.21 |
| regulation of kinase activity | 51 | 1.28 | 1.15 | 1.34 | 1.13 | 1.31 | 1.38 | 1.19 |
| regulation of translation in response to stress | 4 | 2.6 | -1.22 | -1.07 | -1.29 | 1.28 | 1.91 | 1.61 |
| regulation of translational initiation in response to stress | 4 | 2.6 | -1.22 | -1.07 | -1.29 | 1.28 | 1.91 | 1.61 |
| Ku70:Ku80 complex | 1 | 1.33 | 1.54 | 1.93 | 1.1 | 1.72 | 1.13 | 1.74 |
| sequence-specific DNA binding | 271 | 1.14 | 1.03 | 1.1 | 1.01 | 1.08 | 1.09 | 1.12 |
| structure-specific DNA binding | 33 | 1.17 | 1.16 | 1.28 | 1.17 | 1.27 | 1.16 | 1.07 |
| regulation of insulin-like growth factor receptor signaling pathway | 1 | -1.19 | -1.04 | -1.16 | -1.01 | -1.23 | -1.03 | -1.09 |
| maintenance of DNA repeat elements | 2 | 1.62 | -1.0 | 1.14 | 1.16 | 1.41 | 1.05 | -1.1 |
| peroxisomal transport | 3 | 1.5 | 1.17 | -1.6 | 1.44 | 1.46 | -2.19 | -1.64 |
| nuclear replication fork | 11 | 1.14 | 1.11 | 1.17 | 1.2 | 1.33 | 1.04 | -1.23 |
| nuclear replisome | 11 | 1.14 | 1.11 | 1.17 | 1.2 | 1.33 | 1.04 | -1.23 |
| cellular amide metabolic process | 5 | -1.4 | -1.19 | -1.57 | -1.07 | -1.0 | -1.05 | -1.49 |
| amide biosynthetic process | 3 | -2.21 | -1.28 | -2.94 | -1.05 | 1.16 | -1.62 | -1.65 |
| cellular amide catabolic process | 1 | 2.85 | -1.01 | 1.16 | 1.03 | -1.08 | 1.74 | -1.1 |
| regulation of DNA-dependent transcription in response to stress | 2 | -1.52 | 1.4 | 1.63 | 1.11 | 1.53 | 1.64 | 1.52 |
| protein self-association | 5 | -1.05 | 1.16 | 1.37 | 1.04 | 1.23 | 1.34 | 1.34 |
| cellular protein complex assembly | 71 | -1.01 | 1.07 | 1.26 | 1.11 | 1.23 | 1.07 | -1.04 |
| cellular protein complex disassembly | 15 | -1.02 | 1.15 | 1.27 | 1.13 | 1.29 | 1.16 | 1.02 |
| delta DNA polymerase complex | 1 | -1.2 | -1.0 | -1.18 | 1.32 | 1.12 | -1.14 | -1.49 |
| ncRNA 3'-end processing | 2 | 1.0 | 1.06 | 1.29 | 1.13 | 1.22 | 1.14 | 1.21 |
| ncRNA polyadenylation | 1 | -1.36 | -1.04 | -1.09 | 1.02 | -1.09 | -1.15 | 1.04 |
| ncRNA polyadenylation involved in polyadenylation-dependent ncRNA catabolic process | 1 | -1.36 | -1.04 | -1.09 | 1.02 | -1.09 | -1.15 | 1.04 |
| RNA polyadenylation | 17 | 1.35 | 1.11 | 1.23 | 1.12 | 1.29 | 1.65 | 1.19 |
| modification-dependent macromolecule catabolic process | 95 | 1.16 | 1.13 | 1.32 | 1.13 | 1.29 | 1.1 | 1.12 |
| polyadenylation-dependent RNA catabolic process | 1 | -1.36 | -1.04 | -1.09 | 1.02 | -1.09 | -1.15 | 1.04 |
| polyadenylation-dependent ncRNA catabolic process | 1 | -1.36 | -1.04 | -1.09 | 1.02 | -1.09 | -1.15 | 1.04 |
| inositol phosphate metabolic process | 7 | 1.45 | 1.12 | 1.25 | -1.03 | 1.17 | 1.27 | 1.19 |
| dicarboxylic acid metabolic process | 14 | -1.06 | 1.04 | 1.07 | -1.22 | 1.01 | 1.26 | -1.08 |
| engulfment of apoptotic cell | 2 | 1.24 | 1.08 | 1.26 | 1.1 | 1.14 | 1.16 | 1.03 |
| regulation of phosphoprotein phosphatase activity | 3 | 1.09 | 1.05 | 1.0 | 1.06 | 1.03 | 1.05 | -1.09 |
| axon terminus | 26 | 1.26 | 1.07 | 1.08 | -1.15 | 1.13 | 1.67 | 1.27 |
| post-translational protein modification | 2 | -1.4 | 1.08 | 1.03 | 1.29 | 1.32 | -1.28 | -1.35 |
| detection of pheromone | 7 | 1.05 | 1.03 | 1.02 | -1.0 | -1.06 | 1.05 | -1.08 |
| photoreceptor cell fate specification | 11 | 1.11 | 1.04 | 1.07 | 1.17 | 1.1 | -1.16 | -1.1 |
| 2,3-diketo-5-methylthiopentyl-1-phosphate enolase activity | 1 | 1.92 | 1.16 | 1.15 | 1.28 | 1.06 | 2.87 | -1.21 |
| 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase activity | 1 | 1.92 | 1.16 | 1.15 | 1.28 | 1.06 | 2.87 | -1.21 |
| 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity | 1 | 1.67 | 1.48 | 1.35 | 1.44 | 1.31 | 1.18 | 1.35 |
| dihydrolipoyllysine-residue (2-methylpropanoyl)transferase activity | 1 | 1.74 | 1.44 | 1.67 | 1.01 | 1.35 | 1.38 | 1.19 |
| cyclic nucleotide-gated ion channel activity | 3 | -1.19 | 1.0 | 1.03 | -1.02 | 1.02 | -1.02 | -1.19 |
| acireductone synthase activity | 1 | 1.92 | 1.16 | 1.15 | 1.28 | 1.06 | 2.87 | -1.21 |
| regulation of multi-organism process | 51 | 1.44 | -1.06 | 1.09 | -1.01 | 1.09 | 1.15 | 1.1 |
| negative regulation of multi-organism process | 14 | 1.67 | -1.01 | 1.49 | 1.13 | 1.44 | 1.14 | -1.02 |
| positive regulation of multi-organism process | 1 | 8.35 | -1.25 | -1.3 | -1.07 | -1.15 | 1.62 | -1.07 |
| regulation of symbiosis, encompassing mutualism through parasitism | 7 | 2.04 | -1.02 | 1.37 | 1.07 | 1.52 | 1.47 | 1.08 |
| macromolecular complex subunit organization | 187 | 1.08 | 1.11 | 1.28 | 1.14 | 1.27 | 1.13 | 1.04 |
| cellular component maintenance | 23 | -1.02 | 1.04 | 1.3 | 1.14 | 1.3 | -1.03 | 1.0 |
| histone H3 acetylation | 6 | 1.33 | 1.29 | 1.52 | 1.18 | 1.35 | 1.37 | 1.35 |
| histone H4 acetylation | 4 | 1.15 | 1.24 | 1.16 | 1.28 | 1.1 | -1.1 | 1.08 |
| histone H2A acetylation | 1 | 1.8 | 1.23 | 1.54 | 1.26 | 1.37 | 1.4 | 1.76 |
| histone H3-K9 acetylation | 2 | 1.05 | 1.43 | 1.59 | 1.17 | 1.33 | 1.34 | 1.37 |
| histone H4-K12 acetylation | 1 | 1.24 | 1.29 | 1.32 | 1.24 | 1.1 | -1.34 | -1.14 |
| histone H4-R3 methylation | 1 | 1.39 | 1.1 | 1.39 | 1.26 | 1.71 | 1.23 | 1.23 |
| histone H3-S10 phosphorylation | 1 | 1.24 | 1.29 | 1.32 | 1.24 | 1.1 | -1.34 | -1.14 |
| histone acetyltransferase activity (H3-K9 specific) | 1 | -1.13 | 1.58 | 1.91 | 1.11 | 1.62 | 2.4 | 2.13 |
| H2A histone acetyltransferase activity | 1 | 1.8 | 1.23 | 1.54 | 1.26 | 1.37 | 1.4 | 1.76 |
| histone methyltransferase activity (H4-R3 specific) | 1 | 1.39 | 1.1 | 1.39 | 1.26 | 1.71 | 1.23 | 1.23 |
| regulation of DNA methylation | 3 | 1.11 | 1.01 | 1.05 | 1.05 | 1.09 | 1.06 | -1.09 |
| cell wall macromolecule metabolic process | 9 | -1.18 | -1.16 | -1.42 | -1.31 | -1.28 | -2.35 | -1.08 |
| glucan metabolic process | 9 | 2.15 | 1.01 | -1.12 | 1.0 | 1.05 | 2.73 | 1.34 |
| regulation of system process | 57 | 1.06 | 1.07 | 1.16 | -1.08 | 1.02 | 1.2 | 1.1 |
| regulation of digestive system process | 1 | 1.2 | 1.33 | 1.18 | -1.01 | -1.15 | 1.3 | 1.08 |
| regulation of endocrine process | 2 | -1.1 | -1.02 | 1.14 | 1.0 | 1.02 | 1.69 | 1.12 |
| regulation of anion transport | 6 | -1.08 | 1.02 | 1.16 | 1.01 | -1.03 | 1.23 | -1.16 |
| cellular component biogenesis | 506 | 1.2 | 1.07 | 1.25 | 1.07 | 1.24 | 1.23 | 1.11 |
| regulation of cellular component biogenesis | 111 | 1.03 | 1.13 | 1.34 | 1.1 | 1.31 | 1.14 | 1.21 |
| positive regulation of cellular component biogenesis | 10 | -1.05 | 1.15 | 1.36 | 1.13 | 1.37 | 1.23 | 1.36 |
| membrane biogenesis | 3 | 1.33 | 1.11 | 1.68 | -1.36 | 1.02 | 2.57 | 1.24 |
| negative regulation of molecular function | 29 | 1.35 | 1.11 | 1.32 | 1.11 | 1.23 | 1.24 | 1.25 |
| positive regulation of molecular function | 73 | 1.18 | 1.14 | 1.3 | 1.11 | 1.31 | 1.36 | 1.14 |
| cellular amine metabolic process | 179 | -1.01 | 1.03 | -1.09 | -1.01 | 1.01 | 1.13 | -1.01 |
| modulation of growth of symbiont involved in interaction with host | 7 | 2.04 | -1.02 | 1.37 | 1.07 | 1.52 | 1.47 | 1.08 |
| negative regulation of growth of symbiont involved in interaction with host | 7 | 2.04 | -1.02 | 1.37 | 1.07 | 1.52 | 1.47 | 1.08 |
| histone H3-K14 acetylation | 1 | 1.24 | 1.29 | 1.32 | 1.24 | 1.1 | -1.34 | -1.14 |
| 'de novo' UMP biosynthetic process | 1 | 1.61 | 1.06 | 1.09 | 1.15 | 1.38 | -1.09 | 1.01 |
| fully spanning plasma membrane | 1 | -1.09 | 1.03 | 1.58 | 1.01 | 1.26 | -1.16 | 1.18 |
| cellular metabolic process | 2625 | 1.15 | 1.06 | 1.11 | 1.08 | 1.17 | 1.16 | 1.03 |
| primary metabolic process | 3122 | -1.05 | -1.03 | -1.02 | 1.03 | 1.07 | -1.05 | -1.05 |
| cellular lipid catabolic process | 27 | -1.83 | -1.24 | -1.62 | 1.14 | 1.08 | -2.79 | -1.74 |
| regulation of multicellular organismal metabolic process | 1 | 1.86 | -1.04 | 1.59 | -1.1 | 2.08 | 2.71 | 1.26 |
| cellular polysaccharide catabolic process | 1 | 2.98 | -1.08 | 1.5 | -1.31 | 1.12 | 4.86 | 1.33 |
| cellular catabolic process | 329 | 1.02 | 1.05 | 1.09 | 1.07 | 1.15 | 1.03 | 1.01 |
| cellular biosynthetic process | 948 | 1.19 | 1.05 | 1.09 | 1.06 | 1.14 | 1.19 | 1.03 |
| cellular lipid metabolic process | 195 | 1.01 | -1.1 | -1.11 | -1.02 | 1.11 | -1.13 | -1.02 |
| cellular protein catabolic process | 106 | 1.15 | 1.12 | 1.31 | 1.13 | 1.28 | 1.07 | 1.08 |
| cellular macromolecule metabolic process | 1842 | 1.19 | 1.1 | 1.2 | 1.12 | 1.23 | 1.19 | 1.07 |
| cellular carbohydrate metabolic process | 165 | -1.04 | -1.01 | -1.23 | 1.0 | 1.01 | -1.04 | -1.1 |
| cellular polysaccharide metabolic process | 15 | 1.66 | 1.11 | 1.03 | 1.05 | 1.17 | 1.91 | 1.35 |
| cellular macromolecule catabolic process | 146 | 1.19 | 1.11 | 1.29 | 1.12 | 1.29 | 1.13 | 1.13 |
| cellular protein metabolic process | 1120 | 1.17 | 1.09 | 1.19 | 1.1 | 1.21 | 1.2 | 1.06 |
| glycerol ether catabolic process | 2 | 1.6 | 1.03 | -1.14 | -1.01 | 1.9 | -1.45 | -1.11 |
| cellular nitrogen compound catabolic process | 61 | -1.05 | 1.02 | -1.0 | 1.04 | 1.03 | -1.12 | 1.02 |
| cellular nitrogen compound biosynthetic process | 185 | 1.18 | 1.04 | -1.04 | -1.01 | -1.02 | 1.25 | 1.03 |
| sulfur compound biosynthetic process | 25 | 1.09 | 1.17 | 1.2 | 1.11 | 1.05 | 1.23 | 1.23 |
| sulfur compound catabolic process | 1 | 1.48 | 1.28 | 1.32 | 1.05 | 1.07 | 1.16 | -1.01 |
| cellular carbohydrate catabolic process | 39 | 1.53 | 1.02 | -1.21 | -1.06 | -1.05 | 1.4 | 1.2 |
| small molecule metabolic process | 599 | -1.01 | -1.0 | -1.11 | -1.01 | 1.02 | 1.02 | -1.06 |
| small molecule catabolic process | 81 | 1.2 | -1.02 | -1.27 | -1.08 | -1.02 | 1.19 | -1.01 |
| small molecule biosynthetic process | 98 | 1.12 | -1.09 | -1.26 | -1.12 | -1.12 | 1.16 | 1.13 |
| dendrite terminus | 1 | -1.38 | 1.0 | -1.11 | -1.02 | -1.17 | -1.19 | 1.01 |
| axonal growth cone | 3 | 1.35 | 1.17 | 1.49 | 1.21 | 1.37 | 1.33 | 1.2 |
| cell body | 27 | -1.0 | -1.02 | 1.19 | 1.05 | 1.12 | 1.07 | 1.01 |
| neuron projection terminus | 26 | 1.26 | 1.07 | 1.08 | -1.15 | 1.13 | 1.67 | 1.27 |
| neuron spine | 4 | 1.47 | 1.02 | 1.26 | -1.09 | 1.32 | 1.72 | 1.43 |
| wound healing, spreading of cells | 1 | -1.04 | -1.12 | 1.46 | 1.28 | 1.4 | -1.15 | 1.1 |
| regulation of transcription involved in anterior/posterior axis specification | 2 | 1.78 | 1.0 | 1.2 | -1.2 | -1.03 | 1.22 | -1.04 |
| dendritic spine head | 3 | 1.62 | -1.01 | 1.32 | -1.1 | 1.45 | 2.01 | 1.64 |
| sodium-dependent phosphate transport | 1 | -51.3 | 1.56 | -15.93 | -1.24 | 2.78 | -3.27 | -134.18 |
| cellular response to fibroblast growth factor stimulus | 10 | 1.63 | -1.12 | 2.13 | -1.03 | 1.14 | 1.56 | 1.09 |
| small conjugating protein ligase binding | 9 | 1.11 | 1.17 | 1.66 | 1.19 | 1.63 | 1.3 | 1.36 |
| small protein conjugating enzyme binding | 1 | 1.42 | -1.23 | 1.7 | 1.22 | 1.08 | 1.57 | -1.21 |
| ribosomal subunit | 153 | 1.21 | 1.04 | 1.16 | 1.13 | 1.19 | 1.3 | -1.18 |
| interspecies interaction between organisms | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| extracellular matrix part | 13 | 1.55 | -1.17 | 1.15 | -1.23 | 1.13 | 2.04 | 1.39 |
| extracellular region part | 219 | 1.0 | -1.15 | -1.2 | -1.18 | -1.13 | -1.0 | 1.1 |
| organelle part | 1903 | 1.14 | 1.08 | 1.18 | 1.1 | 1.2 | 1.19 | 1.03 |
| virion part | 7 | -1.09 | -1.36 | -1.17 | -1.1 | -1.08 | 1.28 | -1.28 |
| intracellular part | 3721 | 1.14 | 1.07 | 1.15 | 1.08 | 1.18 | 1.16 | 1.05 |
| membrane part | 1459 | 1.01 | -1.02 | 1.01 | -1.03 | 1.01 | 1.09 | 1.01 |
| chromosomal part | 250 | 1.18 | 1.11 | 1.21 | 1.15 | 1.28 | 1.17 | 1.07 |
| nuclear part | 738 | 1.22 | 1.12 | 1.25 | 1.18 | 1.29 | 1.17 | 1.07 |
| mitochondrial part | 343 | 1.07 | 1.07 | 1.15 | 1.07 | 1.12 | 1.42 | -1.24 |
| cytoskeletal part | 502 | 1.11 | 1.03 | 1.14 | 1.03 | 1.2 | 1.16 | 1.06 |
| Golgi apparatus part | 80 | -1.01 | 1.12 | 1.12 | 1.1 | 1.15 | 1.15 | 1.34 |
| endoplasmic reticulum part | 101 | -1.09 | 1.11 | 1.08 | 1.08 | 1.06 | -1.11 | 1.12 |
| cytoplasmic vesicle part | 19 | -1.14 | 1.1 | 1.06 | 1.04 | 1.1 | 1.17 | 1.23 |
| vacuolar part | 30 | -1.55 | -1.0 | 1.28 | -1.14 | -1.08 | 1.41 | -1.12 |
| microbody part | 14 | 1.87 | 1.14 | -1.32 | 1.28 | 1.36 | -1.33 | -1.09 |
| endosomal part | 3 | -1.2 | 1.14 | 1.51 | 1.07 | 1.24 | 1.91 | 1.43 |
| cilium part | 3 | -1.04 | 1.08 | 1.04 | 1.16 | -1.12 | -1.04 | -1.07 |
| microtubule-based flagellum part | 1 | 1.03 | 1.24 | 1.01 | 1.09 | 1.02 | -1.11 | 1.01 |
| cytoplasmic part | 1575 | 1.09 | 1.05 | 1.09 | 1.04 | 1.13 | 1.18 | -1.01 |
| cytosolic part | 124 | 1.25 | 1.04 | 1.13 | 1.06 | 1.16 | 1.14 | -1.01 |
| intracellular organelle part | 1884 | 1.13 | 1.08 | 1.18 | 1.1 | 1.2 | 1.19 | 1.03 |
| axoneme part | 13 | -1.04 | 1.02 | 1.03 | 1.02 | -1.05 | 1.0 | -1.11 |
| cell cortex part | 52 | -1.17 | 1.2 | 1.4 | 1.11 | 1.36 | 1.08 | 1.09 |
| contractile fiber part | 31 | 2.32 | -1.44 | -1.08 | -1.22 | 1.23 | 1.92 | -1.17 |
| microtubule organizing center part | 25 | 1.19 | 1.06 | 1.11 | 1.1 | 1.16 | 1.11 | 1.02 |
| nucleoplasm part | 272 | 1.24 | 1.14 | 1.27 | 1.18 | 1.27 | 1.16 | 1.09 |
| nucleolar part | 9 | 1.24 | 1.15 | 1.22 | 1.3 | 1.26 | 1.11 | 1.02 |
| nuclear membrane part | 2 | -1.02 | -2.05 | 1.59 | 1.21 | 2.03 | -1.95 | -2.17 |
| nuclear chromosome part | 80 | 1.18 | 1.13 | 1.23 | 1.15 | 1.27 | 1.13 | 1.03 |
| mitochondrial membrane part | 108 | 1.14 | 1.02 | 1.21 | 1.03 | 1.09 | 1.55 | -1.26 |
| synapse part | 110 | 1.06 | 1.08 | 1.15 | -1.02 | 1.12 | 1.19 | 1.17 |
| plasma membrane part | 322 | 1.24 | -1.03 | 1.09 | -1.04 | 1.07 | 1.24 | 1.14 |
| flagellum part | 1 | 1.03 | 1.24 | 1.01 | 1.09 | 1.02 | -1.11 | 1.01 |
| external encapsulating structure part | 11 | 1.96 | -1.06 | 1.02 | -1.08 | -1.01 | 1.22 | 1.03 |
| cell projection part | 75 | 1.15 | 1.03 | 1.15 | -1.04 | 1.1 | 1.34 | 1.14 |
| cell part | 4600 | 1.14 | 1.06 | 1.13 | 1.06 | 1.16 | 1.15 | 1.06 |
| secondary metabolite biosynthetic process | 25 | 1.42 | -1.09 | -1.13 | -1.11 | -1.11 | 1.14 | 1.2 |
| double-strand break repair via single-strand annealing | 2 | 1.06 | -1.0 | 1.36 | -1.08 | 1.26 | 1.06 | 1.04 |
| double-strand break repair via synthesis-dependent strand annealing | 5 | 1.1 | 1.19 | 1.07 | 1.18 | 1.1 | -1.15 | 1.06 |
| maintenance of fidelity involved in DNA-dependent DNA replication | 1 | -1.49 | 1.01 | -1.08 | 1.09 | -1.02 | -1.34 | -1.39 |
| glycerolipid biosynthetic process | 31 | -1.11 | -1.12 | 1.05 | -1.1 | 1.14 | 1.03 | -1.04 |
| sensory organ precursor cell division | 8 | -1.01 | 1.07 | 1.27 | 1.11 | 1.3 | -1.03 | -1.04 |
| protein import into mitochondrial inner membrane | 7 | 1.09 | 1.08 | 1.28 | 1.16 | 1.18 | 1.3 | -1.13 |
| protein targeting to ER | 15 | -1.21 | 1.37 | -1.05 | 1.04 | -1.19 | -1.54 | 1.26 |
| regulated secretory pathway | 1 | 4.01 | -1.11 | 1.04 | -1.06 | 1.03 | 1.05 | 1.13 |
| regulation of viral genome replication | 4 | 1.27 | 1.23 | 1.52 | 1.17 | 1.6 | 1.47 | 1.34 |
| negative regulation of viral genome replication | 4 | 1.27 | 1.23 | 1.52 | 1.17 | 1.6 | 1.47 | 1.34 |
| innate immune response | 52 | 1.72 | -1.32 | -1.33 | -1.27 | -1.15 | 1.39 | 1.14 |
| regulation of innate immune response | 11 | 2.21 | -1.14 | 1.08 | -1.08 | 1.02 | 1.67 | 1.13 |
| positive regulation of innate immune response | 5 | 1.98 | -1.06 | 1.18 | -1.06 | 1.11 | 1.79 | 1.28 |
| intermediate filament-based process | 1 | 1.1 | 1.12 | 1.07 | -1.03 | -1.01 | 1.1 | 1.01 |
| intermediate filament cytoskeleton organization | 1 | 1.1 | 1.12 | 1.07 | -1.03 | -1.01 | 1.1 | 1.01 |
| intermediate filament cytoskeleton | 2 | 1.28 | 1.06 | -1.0 | 1.32 | 1.15 | -1.17 | -1.1 |
| membrane raft | 3 | 1.56 | 1.01 | 1.01 | 1.02 | 1.02 | 1.3 | 1.71 |
| meiotic chromosome segregation | 51 | 1.14 | 1.08 | 1.19 | 1.11 | 1.27 | 1.14 | 1.09 |
| uridine-diphosphatase activity | 1 | -1.51 | 1.28 | -1.3 | 1.09 | -1.1 | -3.57 | 1.37 |
| development of primary sexual characteristics | 59 | 1.13 | 1.06 | 1.12 | 1.07 | 1.24 | 1.02 | 1.08 |
| response to copper ion | 3 | -14.42 | 1.56 | 1.38 | 1.09 | -1.89 | -6.59 | -1.3 |
| homologous chromosome segregation | 4 | 1.06 | 1.11 | 1.04 | 1.17 | 1.06 | -1.06 | -1.12 |
| myosin II binding | 2 | -1.98 | 1.16 | 2.07 | 1.03 | 1.58 | -1.28 | 1.01 |
| neuronal ion channel clustering | 1 | 1.29 | -1.71 | -1.84 | -1.91 | -1.32 | 16.38 | -1.61 |
| cell fate commitment | 405 | 1.16 | 1.06 | 1.16 | 1.05 | 1.2 | 1.16 | 1.12 |
| asymmetric protein localization involved in cell fate determination | 14 | -1.35 | 1.16 | 1.3 | 1.08 | 1.48 | 1.01 | 1.07 |
| cell-cell signaling involved in cell fate commitment | 189 | 1.17 | 1.06 | 1.11 | 1.08 | 1.2 | 1.17 | 1.05 |
| fusome | 32 | -1.08 | 1.17 | 1.29 | 1.05 | 1.26 | 1.27 | 1.27 |
| spectrosome | 6 | -1.16 | 1.09 | 1.81 | 1.07 | 1.73 | 1.26 | 1.26 |
| intercellular bridge | 17 | -1.22 | 1.11 | 1.21 | 1.11 | 1.33 | 1.25 | 1.34 |
| germline ring canal | 16 | -1.22 | 1.1 | 1.19 | 1.11 | 1.33 | 1.27 | 1.35 |
| glutathione dehydrogenase (ascorbate) activity | 4 | 1.29 | 1.17 | 1.1 | 1.04 | 1.18 | -1.03 | 1.04 |
| basal protein localization | 6 | -1.45 | 1.22 | 1.54 | 1.08 | 1.45 | -1.08 | -1.07 |
| apical protein localization | 9 | -1.03 | 1.16 | 1.11 | 1.05 | 1.43 | 1.01 | 1.44 |
| apical part of cell | 62 | 1.31 | 1.12 | 1.35 | 1.0 | 1.28 | 1.5 | 1.32 |
| basal part of cell | 11 | -1.08 | 1.22 | 1.48 | 1.16 | 1.48 | 1.32 | 1.21 |
| apical cortex | 27 | -1.0 | 1.23 | 1.46 | 1.07 | 1.36 | 1.19 | 1.13 |
| basal cortex | 7 | -1.21 | 1.31 | 1.34 | 1.19 | 1.4 | -1.05 | 1.17 |
| glutamate synthase activity, NADH or NADPH as acceptor | 1 | 2.14 | 1.19 | 1.24 | 1.03 | -1.73 | 3.68 | 2.51 |
| translation regulator activity | 15 | 1.38 | 1.09 | 1.42 | 1.04 | 1.28 | 1.17 | 1.47 |
| establishment of protein localization | 251 | 1.07 | 1.16 | 1.31 | 1.12 | 1.24 | 1.13 | 1.15 |
| maintenance of protein location | 41 | 1.22 | 1.1 | 1.26 | 1.06 | 1.28 | 1.3 | 1.26 |
| zonula adherens assembly | 8 | -1.0 | 1.29 | 1.57 | 1.09 | 1.44 | 1.45 | 1.58 |
| regulation of circadian sleep/wake cycle, sleep | 13 | -1.21 | 1.24 | 1.19 | -1.03 | 1.06 | 1.12 | 1.26 |
| establishment or maintenance of neuroblast polarity | 10 | -1.07 | 1.12 | 1.77 | 1.0 | 1.45 | 1.54 | 1.19 |
| establishment or maintenance of epithelial cell apical/basal polarity | 23 | 1.37 | 1.08 | 1.39 | -1.01 | 1.36 | 1.42 | 1.26 |
| establishment of epithelial cell apical/basal polarity | 7 | 1.7 | 1.04 | 1.19 | -1.07 | 1.56 | 1.4 | 1.47 |
| maintenance of epithelial cell apical/basal polarity | 4 | 1.21 | 1.16 | 1.51 | 1.18 | 1.38 | 1.16 | 1.05 |
| establishment of neuroblast polarity | 4 | -1.89 | 1.09 | 2.72 | -1.04 | 1.73 | -1.21 | 1.17 |
| maintenance of neuroblast polarity | 1 | 1.14 | -1.24 | 1.17 | -1.53 | 1.18 | 15.9 | 1.17 |
| synapse | 139 | 1.14 | 1.05 | 1.16 | -1.03 | 1.13 | 1.25 | 1.17 |
| postsynaptic membrane | 29 | 1.11 | 1.01 | 1.05 | -1.13 | -1.09 | 1.08 | 1.16 |
| neurotransmitter receptor metabolic process | 3 | 1.15 | 1.01 | -1.1 | -1.21 | 1.3 | 1.34 | -1.13 |
| sarcomere organization | 19 | 2.14 | -1.13 | 1.26 | -1.14 | 1.25 | 1.79 | -1.0 |
| cell-cell junction organization | 45 | 1.62 | -1.06 | 1.35 | -1.1 | 1.25 | 1.89 | 1.32 |
| cell-cell junction maintenance | 2 | -1.01 | 1.25 | 1.94 | 1.07 | 1.78 | 1.99 | 1.84 |
| zonula adherens maintenance | 1 | 3.86 | 1.45 | 2.61 | -1.03 | 2.28 | 4.0 | 3.57 |
| tricarboxylic acid cycle enzyme complex | 10 | 1.04 | -1.01 | 1.14 | -1.12 | -1.04 | 1.51 | -1.27 |
| isocitrate dehydrogenase complex (NAD+) | 1 | -1.07 | 1.16 | -1.05 | -1.13 | 1.15 | 1.71 | 1.06 |
| succinate-CoA ligase complex (GDP-forming) | 3 | 1.16 | -1.04 | -1.07 | -1.04 | 1.11 | 1.22 | -1.42 |
| electron transfer flavoprotein complex | 2 | 1.33 | 1.29 | -1.39 | 1.04 | 1.01 | -1.08 | -1.3 |
| pyruvate dehydrogenase (lipoamide) phosphatase complex | 1 | 1.97 | 1.3 | 1.52 | -1.19 | 1.01 | 1.99 | 1.65 |
| succinate dehydrogenase complex (ubiquinone) | 6 | 1.02 | 1.03 | 1.28 | -1.03 | -1.0 | 1.38 | -1.21 |
| proton-transporting ATP synthase complex | 19 | 1.33 | 1.01 | 1.2 | -1.01 | 1.12 | 1.63 | -1.2 |
| proton-transporting ATP synthase complex, catalytic core F(1) | 9 | 1.58 | -1.04 | 1.09 | -1.06 | 1.03 | 1.78 | -1.26 |
| proton-transporting ATP synthase, central stalk | 1 | 1.03 | 1.03 | 1.17 | 1.01 | 1.11 | 1.58 | -1.38 |
| respiratory chain complex I | 37 | 1.15 | 1.04 | 1.27 | 1.07 | 1.11 | 1.72 | -1.29 |
| respiratory chain complex II | 6 | 1.02 | 1.03 | 1.28 | -1.03 | -1.0 | 1.38 | -1.21 |
| respiratory chain complex III | 11 | -1.13 | 1.03 | 1.1 | 1.04 | -1.01 | 1.3 | -1.48 |
| respiratory chain complex IV | 13 | 1.04 | -1.01 | 1.2 | -1.0 | 1.14 | 1.38 | -1.29 |
| succinate dehydrogenase complex | 6 | 1.02 | 1.03 | 1.28 | -1.03 | -1.0 | 1.38 | -1.21 |
| fumarate reductase complex | 6 | 1.02 | 1.03 | 1.28 | -1.03 | -1.0 | 1.38 | -1.21 |
| luciferin monooxygenase activity | 1 | 2.47 | -1.2 | -1.84 | 1.79 | 2.31 | -2.71 | -1.43 |
| nuclear mRNA cis splicing, via spliceosome | 1 | -1.16 | -1.03 | -1.07 | 1.26 | 1.12 | -1.31 | -1.69 |
| alpha-catenin binding | 1 | 1.17 | 1.11 | 1.06 | 1.15 | 1.09 | 1.09 | 1.68 |
| cadherin binding | 2 | 1.22 | -1.01 | 1.46 | 1.06 | 1.25 | 1.4 | 1.02 |
| post-mating behavior | 20 | 1.01 | -1.06 | -1.13 | -1.12 | -1.11 | -1.0 | -1.08 |
| tubulin complex | 10 | -1.16 | 1.02 | 1.08 | 1.02 | 1.1 | -1.0 | -1.09 |
| protein phosphorylated amino acid binding | 1 | -1.42 | 1.22 | 1.22 | 1.19 | 1.41 | -1.04 | -1.04 |
| rhabdomere membrane biogenesis | 3 | 1.33 | 1.11 | 1.68 | -1.36 | 1.02 | 2.57 | 1.24 |
| regulation of compound eye photoreceptor development | 10 | 1.04 | 1.09 | 1.37 | -1.02 | 1.29 | 1.07 | 1.24 |
| positive regulation of compound eye photoreceptor development | 1 | 2.02 | 1.25 | 1.56 | 1.13 | 1.49 | 2.5 | 1.69 |
| negative regulation of compound eye photoreceptor development | 3 | -1.04 | -1.04 | 1.09 | -1.03 | 1.27 | -1.29 | 1.13 |
| equator specification | 6 | -1.25 | -1.43 | -1.32 | -1.53 | -1.4 | -1.25 | 3.26 |
| leukocyte activation | 1 | 2.18 | -2.11 | 2.19 | 1.49 | 1.48 | 1.37 | 1.38 |
| carnitine biosynthetic process | 1 | 1.3 | -1.08 | -1.07 | -1.13 | -1.19 | 1.96 | -1.06 |
| cellular respiration | 97 | 1.02 | 1.04 | 1.22 | 1.01 | 1.13 | 1.55 | -1.22 |
| clathrin-coated endocytic vesicle | 1 | -1.65 | 1.56 | 1.35 | 1.0 | 1.12 | 1.4 | 1.32 |
| farnesyl diphosphate biosynthetic process | 1 | 4.21 | 1.14 | 2.47 | -1.3 | 1.39 | 2.06 | 1.87 |
| farnesyl diphosphate metabolic process | 1 | 4.21 | 1.14 | 2.47 | -1.3 | 1.39 | 2.06 | 1.87 |
| quinone cofactor biosynthetic process | 5 | 1.2 | 1.14 | 1.21 | 1.14 | 1.17 | 1.39 | 1.02 |
| regulation of nitric oxide biosynthetic process | 1 | 1.39 | -1.03 | 1.89 | -1.0 | 1.66 | 2.5 | 1.68 |
| positive regulation of nitric oxide biosynthetic process | 1 | 1.39 | -1.03 | 1.89 | -1.0 | 1.66 | 2.5 | 1.68 |
| male courtship behavior, veined wing generated song production | 12 | 1.05 | 1.04 | 1.45 | 1.15 | 1.09 | -1.02 | 1.01 |
| negative regulation of female receptivity, post-mating | 8 | -1.24 | 1.06 | 1.02 | 1.03 | 1.16 | -1.13 | -1.08 |
| myoblast differentiation | 1 | -1.07 | -1.09 | 1.09 | -1.1 | 1.09 | -1.05 | 1.02 |
| endothelial cell differentiation | 3 | 1.7 | 1.01 | 1.41 | -1.21 | 1.32 | 4.52 | 2.14 |
| mitotic cell cycle, embryonic | 23 | 1.34 | 1.02 | 1.19 | -1.03 | 1.26 | 1.27 | 1.41 |
| bicoid mRNA localization | 8 | 1.29 | 1.08 | 1.35 | 1.13 | 1.31 | 1.35 | 1.11 |
| pole plasm oskar mRNA localization | 30 | -1.04 | 1.2 | 1.34 | 1.17 | 1.3 | 1.21 | 1.16 |
| ecdysteroid metabolic process | 15 | 1.26 | -1.14 | -1.0 | -1.06 | 1.15 | 1.51 | 1.09 |
| ecdysteroid biosynthetic process | 12 | 1.31 | -1.17 | -1.03 | -1.07 | 1.06 | 1.52 | 1.13 |
| R8 cell development | 3 | 1.3 | 1.12 | 1.37 | 1.13 | 1.55 | 1.07 | 1.27 |
| R8 cell fate specification | 9 | 1.18 | 1.04 | 1.11 | 1.19 | 1.18 | -1.18 | -1.1 |
| R8 cell differentiation | 17 | 1.18 | 1.05 | 1.21 | 1.12 | 1.31 | 1.03 | 1.07 |
| R7 cell differentiation | 36 | 1.32 | 1.13 | 1.36 | 1.07 | 1.37 | 1.31 | 1.33 |
| R7 cell development | 16 | 1.23 | 1.15 | 1.34 | 1.07 | 1.33 | 1.14 | 1.28 |
| regulation of R8 cell spacing in compound eye | 5 | 1.17 | 1.07 | 1.1 | 1.16 | 1.41 | 1.4 | 1.18 |
| R8 cell-mediated photoreceptor organization | 2 | 1.29 | 1.14 | 1.27 | 1.19 | 1.33 | 1.03 | 1.2 |
| response to ethanol | 44 | 1.51 | 1.01 | 1.21 | -1.06 | 1.21 | 1.25 | 1.3 |
| response to ether | 3 | -1.07 | 1.05 | -1.12 | -1.04 | -1.19 | 1.04 | -1.17 |
| behavioral response to ethanol | 35 | 1.52 | -1.02 | 1.16 | -1.06 | 1.28 | 1.26 | 1.37 |
| behavioral response to ether | 2 | -1.03 | 1.05 | -1.07 | -1.06 | -1.12 | 1.18 | -1.13 |
| locomotor rhythm | 31 | 1.06 | 1.1 | 1.36 | -1.15 | -1.02 | 1.63 | 1.1 |
| nurse cell apoptotic process | 13 | 1.39 | -1.0 | 1.22 | -1.05 | 1.04 | 1.07 | 1.25 |
| regulation of nurse cell apoptotic process | 5 | 1.15 | 1.23 | 1.79 | 1.07 | 1.35 | 1.85 | 1.46 |
| fusome organization | 12 | 1.01 | 1.08 | 1.22 | 1.09 | 1.28 | 1.3 | 1.26 |
| vesicle targeting to fusome | 1 | -1.29 | -1.05 | -1.05 | 1.11 | -1.02 | -1.43 | 1.36 |
| photoreceptor cell maintenance | 13 | 1.14 | -1.01 | 1.16 | 1.18 | 1.21 | -1.08 | 1.04 |
| pole plasm | 19 | 1.4 | 1.04 | 1.1 | 1.08 | 1.12 | 1.3 | -1.03 |
| male analia development | 2 | 2.83 | 1.39 | 1.21 | 1.02 | 1.24 | 1.39 | 1.47 |
| female analia development | 2 | 2.83 | 1.39 | 1.21 | 1.02 | 1.24 | 1.39 | 1.47 |
| sex comb development | 10 | 1.42 | 1.1 | 1.34 | 1.21 | 1.36 | 1.37 | 1.07 |
| sevenless signaling pathway | 9 | 1.54 | 1.11 | 1.39 | 1.09 | 1.39 | 1.67 | 1.31 |
| regulation of sevenless signaling pathway | 1 | -4.89 | -1.17 | -1.75 | -1.1 | -1.11 | -9.33 | -1.59 |
| dynein binding | 7 | 1.02 | 1.14 | 1.3 | 1.05 | 1.23 | 1.2 | 1.13 |
| dynein light chain binding | 1 | -1.29 | 1.22 | 1.62 | 1.06 | 1.22 | 1.25 | 1.14 |
| dynein intermediate chain binding | 2 | 1.32 | -1.03 | 1.69 | 1.08 | 1.5 | 1.12 | -1.03 |
| gibberellin 20-oxidase activity | 1 | -2.43 | -1.01 | 2.24 | -1.36 | -1.68 | -4.63 | 2.94 |
| regulation of imaginal disc growth | 19 | 1.56 | 1.12 | 1.34 | 1.06 | 1.25 | 1.29 | 1.24 |
| negative regulation of imaginal disc growth | 3 | 1.85 | 1.08 | 1.39 | -1.08 | 1.25 | 1.14 | 1.32 |
| positive regulation of imaginal disc growth | 5 | 1.2 | 1.16 | 1.11 | 1.17 | 1.32 | 1.36 | 1.13 |
| regulation of cell differentiation | 237 | 1.12 | 1.11 | 1.33 | 1.07 | 1.29 | 1.21 | 1.19 |
| negative regulation of cell differentiation | 96 | 1.06 | 1.11 | 1.28 | 1.08 | 1.32 | 1.14 | 1.21 |
| positive regulation of cell differentiation | 16 | 1.4 | 1.12 | 1.39 | 1.03 | 1.32 | 1.44 | 1.39 |
| regulation of hemocyte differentiation | 22 | 1.14 | 1.14 | 1.17 | 1.04 | 1.25 | 1.43 | 1.49 |
| negative regulation of hemocyte differentiation | 8 | 1.16 | 1.08 | 1.01 | -1.03 | 1.06 | 1.37 | 1.55 |
| positive regulation of hemocyte differentiation | 1 | -1.18 | -1.04 | -1.07 | -1.26 | 1.04 | 1.06 | 5.07 |
| regulation of neuron differentiation | 76 | 1.07 | 1.12 | 1.36 | 1.07 | 1.33 | 1.17 | 1.17 |
| negative regulation of neuron differentiation | 5 | -1.22 | 1.17 | 1.2 | 1.05 | 1.31 | -1.53 | 1.09 |
| positive regulation of neuron differentiation | 8 | 1.3 | 1.17 | 1.47 | 1.09 | 1.39 | 1.19 | 1.25 |
| regulation of R7 cell differentiation | 11 | 1.0 | 1.13 | 1.24 | 1.05 | 1.3 | -1.16 | 1.17 |
| negative regulation of R7 cell differentiation | 1 | -4.89 | -1.17 | -1.75 | -1.1 | -1.11 | -9.33 | -1.59 |
| positive regulation of R7 cell differentiation | 5 | 1.49 | 1.15 | 1.41 | 1.05 | 1.37 | 1.23 | 1.34 |
| regulation of R8 cell differentiation | 1 | 1.01 | 1.09 | 1.11 | -1.09 | -1.01 | 1.02 | -1.12 |
| negative regulation of R8 cell differentiation | 1 | 1.01 | 1.09 | 1.11 | -1.09 | -1.01 | 1.02 | -1.12 |
| regulation of glial cell differentiation | 1 | 1.08 | 1.07 | 1.03 | 1.0 | -1.04 | -1.0 | -1.04 |
| positive regulation of glial cell differentiation | 1 | 1.08 | 1.07 | 1.03 | 1.0 | -1.04 | -1.0 | -1.04 |
| regulation of salivary gland boundary specification | 8 | 1.56 | -1.2 | -1.1 | 1.04 | 1.14 | 1.2 | -1.02 |
| negative regulation of salivary gland boundary specification | 8 | 1.56 | -1.2 | -1.1 | 1.04 | 1.14 | 1.2 | -1.02 |
| positive regulation of protein catabolic process | 1 | 2.44 | 1.03 | 1.21 | 1.16 | 1.33 | 1.95 | 1.23 |
| negative regulation of cyclin-dependent protein kinase activity | 1 | 2.05 | 1.25 | 2.19 | 1.12 | 2.03 | 2.13 | 2.15 |
| positive regulation of DNA replication | 4 | 1.13 | 1.17 | 1.08 | 1.14 | 1.28 | -1.01 | 1.07 |
| positive regulation of epidermal growth factor receptor signaling pathway | 8 | 1.07 | 1.11 | 1.54 | 1.04 | 1.59 | 1.25 | 1.43 |
| negative regulation of Notch signaling pathway | 22 | 1.04 | 1.15 | 1.34 | 1.08 | 1.39 | 1.3 | 1.33 |
| positive regulation of Notch signaling pathway | 18 | 1.04 | 1.21 | 1.35 | 1.11 | 1.43 | 1.18 | 1.26 |
| positive regulation of S phase of mitotic cell cycle | 5 | 1.47 | 1.12 | 1.21 | 1.09 | 1.33 | 1.59 | 1.23 |
| negative regulation of Toll signaling pathway | 3 | 5.73 | -1.65 | -1.56 | -1.86 | -1.27 | 7.14 | 1.48 |
| positive regulation of Toll signaling pathway | 8 | 1.13 | -1.26 | -1.25 | -1.26 | -1.11 | -1.13 | 1.2 |
| regulation of adenylate cyclase activity | 7 | 1.19 | 1.06 | 1.12 | -1.03 | 1.03 | 1.11 | -1.03 |
| positive regulation of adenylate cyclase activity | 6 | 1.16 | 1.07 | 1.14 | -1.06 | 1.03 | 1.1 | -1.03 |
| regulation of anti-apoptosis | 2 | 1.05 | 1.08 | 1.9 | 1.35 | 1.37 | -1.31 | 1.1 |
| positive regulation of cell adhesion | 2 | 1.24 | 1.07 | 1.55 | 1.24 | 2.26 | 1.04 | 1.52 |
| negative regulation of cell cycle | 44 | 1.23 | 1.15 | 1.3 | 1.13 | 1.41 | 1.3 | 1.26 |
| positive regulation of cell cycle | 19 | 1.15 | 1.12 | 1.16 | 1.12 | 1.4 | 1.36 | 1.11 |
| negative regulation of cell size | 13 | 1.4 | 1.18 | 1.18 | 1.0 | 1.22 | 1.64 | 1.25 |
| positive regulation of cell size | 21 | 1.09 | 1.23 | 1.41 | 1.08 | 1.42 | 1.39 | 1.39 |
| negative regulation of chromatin assembly or disassembly | 1 | 1.22 | 1.33 | 2.18 | 1.07 | 1.72 | 2.05 | 1.61 |
| positive regulation of chromatin assembly or disassembly | 2 | 1.21 | 1.02 | 1.16 | 1.13 | 1.43 | 1.09 | -1.08 |
| negative regulation of eclosion | 1 | -1.18 | 1.19 | 1.16 | 1.18 | 1.03 | -1.13 | 1.04 |
| positive regulation of eclosion | 1 | 1.76 | 1.07 | 1.13 | 1.08 | 1.49 | 1.28 | 1.15 |
| negative regulation of endocytosis | 1 | -1.04 | -1.14 | 1.37 | -1.01 | 1.09 | 1.23 | 1.09 |
| positive regulation of endocytosis | 9 | -1.04 | 1.12 | 1.4 | 1.06 | 1.19 | 1.37 | 1.21 |
| negative regulation of gene expression, epigenetic | 41 | 1.28 | 1.14 | 1.29 | 1.12 | 1.31 | 1.29 | 1.16 |
| positive regulation of gene expression, epigenetic | 14 | 1.54 | 1.18 | 1.37 | 1.13 | 1.35 | 1.46 | 1.35 |
| negative regulation of heart contraction | 2 | 1.15 | -1.01 | -1.01 | 1.01 | -1.02 | 1.08 | -1.02 |
| positive regulation of heart contraction | 1 | -1.08 | 1.01 | 1.03 | 1.02 | -1.06 | -1.21 | -1.22 |
| negative regulation of innate immune response | 3 | 3.2 | -1.5 | -1.25 | -1.29 | -1.32 | 1.18 | 1.06 |
| negative regulation of isoprenoid metabolic process | 1 | -84.21 | -3.59 | 1.42 | 2.07 | 1.14 | -58.36 | -15.07 |
| negative regulation of lipid metabolic process | 2 | -18.13 | -1.6 | 1.31 | 1.68 | 1.16 | -11.26 | -4.64 |
| positive regulation of lipid metabolic process | 1 | -3.9 | 1.39 | 1.21 | 1.36 | 1.18 | -2.17 | -1.43 |
| negative regulation of meiosis | 1 | 1.5 | 1.02 | 1.68 | -1.05 | 1.78 | 1.86 | 1.49 |
| positive regulation of meiosis | 1 | -1.51 | 1.58 | 1.98 | 1.08 | 1.74 | 1.62 | 1.22 |
| negative regulation of mitosis | 15 | 1.05 | 1.11 | 1.17 | 1.19 | 1.35 | 1.07 | 1.0 |
| positive regulation of mitosis | 2 | -1.01 | 1.03 | 1.05 | 1.11 | 1.69 | -1.0 | -1.02 |
| negative regulation of mitotic metaphase/anaphase transition | 12 | 1.08 | 1.1 | 1.18 | 1.19 | 1.38 | 1.04 | 1.01 |
| positive regulation of mitotic metaphase/anaphase transition | 2 | -1.01 | 1.03 | 1.05 | 1.11 | 1.69 | -1.0 | -1.02 |
| negative regulation of striated muscle tissue development | 1 | -1.17 | -1.03 | -1.15 | -1.16 | -1.43 | -1.09 | -1.47 |
| negative regulation of nurse cell apoptotic process | 1 | 1.41 | 1.35 | 1.47 | 1.02 | 1.54 | 2.41 | 1.84 |
| positive regulation of nurse cell apoptotic process | 2 | 1.07 | 1.35 | 1.52 | 1.17 | 1.22 | 1.39 | 1.44 |
| pH reduction | 6 | -1.56 | 1.28 | 1.81 | 1.0 | 1.29 | 1.81 | 1.32 |
| positive regulation of pole plasm oskar mRNA localization | 2 | -1.78 | 1.46 | 1.88 | 1.15 | 2.04 | -1.01 | 1.92 |
| regulation of protein kinase activity | 51 | 1.28 | 1.15 | 1.34 | 1.13 | 1.31 | 1.38 | 1.19 |
| positive regulation of protein kinase activity | 25 | 1.18 | 1.14 | 1.29 | 1.1 | 1.23 | 1.49 | 1.19 |
| negative regulation of proteolysis | 12 | 1.33 | -1.03 | 1.04 | -1.01 | 1.08 | 1.15 | 1.19 |
| positive regulation of proteolysis | 3 | -1.32 | 1.1 | 1.33 | 1.18 | 1.24 | -1.5 | 1.07 |
| negative regulation of sevenless signaling pathway | 1 | -4.89 | -1.17 | -1.75 | -1.1 | -1.11 | -9.33 | -1.59 |
| negative regulation of smoothened signaling pathway | 15 | 1.43 | 1.17 | 1.37 | 1.01 | 1.27 | 1.9 | 1.34 |
| positive regulation of smoothened signaling pathway | 9 | 1.35 | 1.08 | 1.43 | 1.16 | 1.54 | 1.59 | 1.09 |
| negative regulation of synaptic growth at neuromuscular junction | 12 | -1.17 | 1.11 | 1.26 | 1.17 | 1.16 | -1.1 | 1.03 |
| positive regulation of synaptic growth at neuromuscular junction | 7 | -1.19 | 1.15 | 1.24 | 1.09 | 1.31 | 1.0 | 1.41 |
| regulation of transcription during mitosis | 4 | 1.21 | 1.13 | 1.22 | 1.16 | 1.17 | 1.14 | 1.08 |
| regulation of RNA polymerase II transcriptional preinitiation complex assembly | 3 | 1.71 | 1.26 | 1.41 | 1.3 | 1.44 | 1.38 | 1.2 |
| negative regulation of translational elongation | 2 | 1.41 | 1.28 | 1.1 | 1.19 | -1.04 | -1.13 | 1.17 |
| positive regulation of translational elongation | 1 | 1.71 | 1.01 | 1.01 | -1.13 | 1.21 | 1.32 | 1.15 |
| positive regulation of translational termination | 1 | 1.71 | 1.01 | 1.01 | -1.13 | 1.21 | 1.32 | 1.15 |
| negative regulation of DNA recombination | 2 | -1.59 | 1.15 | 1.13 | 1.19 | 1.42 | -1.74 | -1.31 |
| negative regulation of exocytosis | 1 | 1.24 | 1.25 | 1.11 | 1.13 | 1.1 | 1.34 | 1.26 |
| positive regulation of exocytosis | 4 | -1.5 | 1.3 | 1.55 | 1.16 | 1.38 | 1.25 | 1.13 |
| regulation of female receptivity | 18 | 1.06 | -1.2 | -1.2 | -1.22 | -1.06 | 1.34 | 1.14 |
| negative regulation of growth | 41 | 1.29 | 1.12 | 1.35 | 1.07 | 1.32 | 1.34 | 1.19 |
| positive regulation of growth | 42 | 1.0 | 1.19 | 1.31 | 1.08 | 1.32 | 1.25 | 1.31 |
| negative regulation of juvenile hormone metabolic process | 1 | -84.21 | -3.59 | 1.42 | 2.07 | 1.14 | -58.36 | -15.07 |
| negative regulation of mitotic cell cycle | 20 | 1.24 | 1.15 | 1.29 | 1.14 | 1.39 | 1.35 | 1.29 |
| positive regulation of mitotic cell cycle | 13 | 1.23 | 1.16 | 1.25 | 1.11 | 1.43 | 1.47 | 1.21 |
| negative regulation of muscle contraction | 1 | 13.14 | -1.35 | -1.29 | -1.22 | -1.17 | -1.49 | -1.27 |
| negative regulation of nucleobase-containing compound metabolic process | 199 | 1.16 | 1.09 | 1.16 | 1.05 | 1.22 | 1.21 | 1.14 |
| negative regulation of phosphate metabolic process | 8 | 1.4 | 1.03 | 1.23 | 1.05 | 1.2 | 1.33 | 1.24 |
| positive regulation of phosphate metabolic process | 8 | 1.51 | 1.21 | 1.82 | 1.05 | 1.45 | 2.01 | 1.56 |
| positive regulation of circadian sleep/wake cycle, sleep | 2 | 1.24 | 1.16 | 1.04 | -1.02 | 1.05 | 1.25 | 1.29 |
| positive regulation of transcription from RNA polymerase II promoter | 148 | 1.13 | 1.01 | 1.12 | 1.01 | 1.1 | 1.18 | 1.12 |
| negative regulation of nucleotide metabolic process | 2 | 1.18 | 1.06 | 1.18 | 1.14 | 1.04 | 1.11 | -1.15 |
| positive regulation of nucleotide metabolic process | 6 | 1.16 | 1.07 | 1.14 | -1.06 | 1.03 | 1.1 | -1.03 |
| regulation of embryonic development | 64 | 1.08 | 1.16 | 1.34 | 1.09 | 1.37 | 1.37 | 1.21 |
| regulation of female receptivity, post-mating | 14 | 1.03 | -1.09 | -1.21 | -1.16 | -1.11 | 1.05 | -1.06 |
| regulation of oskar mRNA translation | 12 | 1.38 | 1.02 | 1.27 | -1.05 | 1.15 | 1.71 | 1.06 |
| positive regulation of oskar mRNA translation | 3 | 1.65 | 1.1 | 1.35 | -1.02 | 1.27 | 1.67 | 1.22 |
| regulation of transcription from RNA polymerase II promoter, mitotic | 4 | 1.21 | 1.13 | 1.22 | 1.16 | 1.17 | 1.14 | 1.08 |
| ADP metabolic process | 2 | -1.29 | -1.01 | -1.04 | 1.37 | 1.42 | -2.05 | -1.57 |
| ATP metabolic process | 36 | 1.35 | -1.02 | 1.24 | -1.12 | 1.07 | 1.65 | -1.08 |
| CTP metabolic process | 4 | 1.0 | 1.09 | 1.36 | 1.22 | 1.36 | 1.06 | -1.14 |
| GMP metabolic process | 2 | 1.15 | 1.21 | 1.23 | -1.01 | 1.16 | 1.11 | 2.05 |
| GTP metabolic process | 41 | 1.15 | 1.04 | 1.16 | 1.09 | 1.14 | 1.08 | -1.01 |
| IMP metabolic process | 7 | 1.14 | 1.03 | -1.12 | -1.04 | -1.13 | 1.95 | -1.01 |
| TMP metabolic process | 1 | 1.02 | 1.09 | 1.32 | 1.32 | 1.3 | -1.29 | -1.13 |
| UMP metabolic process | 3 | 1.1 | 1.3 | 1.05 | 1.25 | 1.1 | -1.22 | -1.06 |
| UTP metabolic process | 4 | 1.0 | 1.09 | 1.36 | 1.22 | 1.36 | 1.06 | -1.14 |
| cGMP metabolic process | 11 | -1.12 | 1.01 | -1.02 | -1.0 | -1.04 | -1.01 | -1.06 |
| WW domain binding | 3 | 2.14 | -1.08 | 1.3 | -1.28 | -1.29 | 2.35 | 1.56 |
| dTDP metabolic process | 1 | 1.27 | 1.13 | 1.91 | 1.21 | 1.41 | -1.17 | -1.08 |
| dTMP metabolic process | 2 | 1.05 | 1.03 | -1.1 | 1.26 | 1.31 | 1.33 | -1.09 |
| dUTP metabolic process | 1 | -1.36 | -1.27 | 1.07 | 1.7 | 1.82 | -3.47 | -2.13 |
| adenine metabolic process | 1 | 1.17 | 1.37 | 1.23 | 1.24 | 1.3 | 1.33 | -1.5 |
| adenine biosynthetic process | 1 | 1.17 | 1.37 | 1.23 | 1.24 | 1.3 | 1.33 | -1.5 |
| cytidine metabolic process | 3 | -2.8 | 1.04 | -1.39 | 1.2 | 1.32 | -1.87 | -1.06 |
| nucleobase biosynthetic process | 10 | 1.45 | 1.05 | 1.03 | 1.0 | -1.04 | 1.28 | -1.0 |
| nucleobase catabolic process | 3 | 2.92 | -1.42 | -1.24 | -1.35 | -1.35 | 1.45 | 1.16 |
| guanosine biosynthetic process | 2 | -1.08 | 1.07 | 1.01 | 1.12 | 1.08 | -1.67 | -1.22 |
| queuosine metabolic process | 2 | -1.08 | 1.07 | 1.01 | 1.12 | 1.08 | -1.67 | -1.22 |
| 7-methylguanosine biosynthetic process | 2 | -1.08 | 1.07 | 1.01 | 1.12 | 1.08 | -1.67 | -1.22 |
| purine ribonucleoside metabolic process | 12 | 1.18 | 1.15 | -1.0 | 1.18 | 1.26 | -1.09 | -1.09 |
| purine ribonucleoside biosynthetic process | 7 | 1.07 | 1.09 | -1.03 | 1.13 | 1.19 | -1.05 | -1.12 |
| pyrimidine ribonucleoside metabolic process | 10 | -1.32 | 1.14 | 1.04 | 1.22 | 1.27 | -1.25 | -1.09 |
| pyrimidine ribonucleoside catabolic process | 3 | -2.8 | 1.04 | -1.39 | 1.2 | 1.32 | -1.87 | -1.06 |
| pyrimidine nucleoside catabolic process | 3 | -2.8 | 1.04 | -1.39 | 1.2 | 1.32 | -1.87 | -1.06 |
| tetrahydrobiopterin metabolic process | 4 | 7.84 | -1.48 | -1.41 | -1.58 | -1.23 | 1.76 | 2.48 |
| pigment biosynthetic process | 48 | 1.24 | -1.02 | -1.11 | 1.1 | 1.0 | 1.05 | -1.07 |
| ommochrome metabolic process | 18 | 1.35 | 1.13 | 1.12 | 1.06 | 1.06 | 1.08 | 1.16 |
| rhodopsin metabolic process | 9 | -1.72 | -1.52 | -1.96 | 1.48 | 1.03 | -1.74 | -1.95 |
| ocellus pigment metabolic process | 18 | 1.35 | 1.13 | 1.12 | 1.06 | 1.06 | 1.08 | 1.16 |
| heme a metabolic process | 1 | -1.08 | 1.29 | 1.99 | 1.12 | 1.3 | 1.57 | -1.3 |
| alcohol catabolic process | 39 | 1.48 | 1.03 | -1.37 | -1.1 | -1.08 | 1.21 | 1.22 |
| alcohol biosynthetic process | 14 | 1.09 | -1.23 | -1.05 | -1.2 | 1.02 | 1.19 | 1.3 |
| glycerol-3-phosphate catabolic process | 2 | 1.2 | 1.12 | -1.49 | 1.16 | 1.79 | -1.21 | -1.08 |
| polyol biosynthetic process | 1 | 1.86 | -1.46 | -1.12 | -1.51 | 1.13 | 1.83 | 1.71 |
| polyol catabolic process | 3 | 1.78 | -1.48 | -2.01 | -1.34 | 1.14 | 1.58 | -1.45 |
| aldehyde catabolic process | 1 | 1.1 | -1.81 | -2.08 | -1.81 | -1.6 | 3.21 | -1.83 |
| phenol-containing compound biosynthetic process | 1 | 1.3 | 1.0 | -1.05 | -1.09 | 1.04 | -1.06 | -1.0 |
| spermidine catabolic process | 1 | -1.3 | 1.12 | 1.08 | 1.3 | 1.03 | -1.14 | -1.27 |
| nitric oxide metabolic process | 1 | 1.08 | 1.06 | 1.02 | 1.1 | 1.05 | -1.07 | -1.02 |
| indolalkylamine catabolic process | 3 | -1.86 | 1.07 | -1.41 | -1.17 | -1.45 | -1.76 | -1.05 |
| indolalkylamine biosynthetic process | 3 | 1.22 | -1.67 | -1.93 | -1.69 | -1.72 | 1.14 | 2.21 |
| regulation of JNK cascade | 33 | -1.0 | 1.2 | 1.31 | 1.13 | 1.3 | 1.36 | 1.17 |
| negative regulation of JNK cascade | 12 | 1.09 | 1.13 | 1.44 | 1.11 | 1.23 | 1.18 | 1.08 |
| positive regulation of JNK cascade | 12 | 1.07 | 1.28 | 1.47 | 1.07 | 1.29 | 1.52 | 1.25 |
| lateral inhibition | 189 | 1.17 | 1.06 | 1.11 | 1.08 | 1.2 | 1.17 | 1.05 |
| SMAD binding | 2 | 1.44 | -1.05 | -1.06 | -1.04 | -1.05 | -1.04 | 1.16 |
| octopamine metabolic process | 2 | -2.63 | 1.33 | -1.47 | -1.06 | -1.42 | -3.88 | 1.1 |
| octopamine catabolic process | 1 | -8.98 | 1.77 | -2.05 | -1.02 | -2.08 | -14.22 | 1.22 |
| ethanolamine biosynthetic process | 1 | 1.07 | -1.05 | 2.34 | 1.23 | 1.45 | 3.99 | 1.1 |
| phosphatidylethanolamine metabolic process | 1 | 1.07 | -1.05 | 2.34 | 1.23 | 1.45 | 3.99 | 1.1 |
| diacylglycerol metabolic process | 3 | 1.83 | -1.12 | 1.21 | -1.09 | 1.79 | -1.71 | -1.19 |
| CDP-diacylglycerol metabolic process | 1 | 3.07 | 1.1 | 2.07 | -1.42 | 3.13 | -1.11 | 1.4 |
| ecdysteroid catabolic process | 1 | 1.07 | -1.12 | 1.15 | -1.13 | -1.09 | 1.14 | -1.07 |
| amino sugar biosynthetic process | 1 | 1.0 | -1.04 | -1.09 | -1.07 | -1.02 | 1.03 | -1.14 |
| disaccharide biosynthetic process | 3 | 3.57 | -2.83 | -2.37 | -2.36 | -1.16 | 5.45 | -1.46 |
| acetyl-CoA catabolic process | 36 | -1.0 | 1.09 | 1.26 | -1.05 | 1.07 | 1.52 | -1.04 |
| monosaccharide biosynthetic process | 8 | -1.12 | -1.06 | -1.04 | -1.1 | 1.15 | 1.01 | -1.02 |
| monosaccharide catabolic process | 33 | 1.52 | 1.06 | -1.18 | -1.03 | -1.06 | 1.37 | 1.2 |
| GDP-L-fucose metabolic process | 2 | -1.67 | 1.21 | 1.13 | 1.24 | 1.36 | -1.89 | -1.14 |
| N-acetylneuraminate biosynthetic process | 1 | 1.0 | -1.04 | -1.09 | -1.07 | -1.02 | 1.03 | -1.14 |
| CMP-N-acetylneuraminate metabolic process | 1 | 1.0 | -1.04 | -1.09 | -1.07 | -1.02 | 1.03 | -1.14 |
| carboxylic acid biosynthetic process | 65 | 1.05 | -1.1 | -1.35 | -1.11 | -1.17 | 1.1 | 1.09 |
| carboxylic acid catabolic process | 40 | 1.0 | -1.07 | -1.2 | -1.06 | 1.05 | 1.14 | -1.23 |
| D-amino acid metabolic process | 1 | 1.5 | 1.05 | 1.18 | 1.45 | -1.04 | 1.27 | -1.33 |
| regulation of JAK-STAT cascade | 17 | -1.03 | 1.16 | 1.32 | 1.08 | 1.3 | 1.09 | 1.25 |
| negative regulation of JAK-STAT cascade | 5 | -1.37 | 1.15 | 1.03 | 1.03 | 1.17 | -1.54 | 1.05 |
| positive regulation of JAK-STAT cascade | 3 | -1.1 | 1.05 | 1.09 | 1.11 | 1.21 | -1.07 | 1.08 |
| organophosphate catabolic process | 4 | 2.25 | 1.02 | 1.6 | 1.03 | 1.46 | 1.59 | 1.17 |
| L-cysteine metabolic process | 1 | 1.24 | 1.05 | 1.09 | -1.01 | -1.04 | 1.21 | 1.11 |
| icosanoid biosynthetic process | 1 | -1.74 | 1.11 | -2.01 | 1.51 | 1.27 | -1.87 | -1.55 |
| neutral lipid biosynthetic process | 3 | 2.65 | 1.06 | 2.5 | -1.19 | 1.69 | 1.64 | 1.56 |
| neutral lipid catabolic process | 2 | 1.6 | 1.03 | -1.14 | -1.01 | 1.9 | -1.45 | -1.11 |
| acylglycerol biosynthetic process | 3 | 2.65 | 1.06 | 2.5 | -1.19 | 1.69 | 1.64 | 1.56 |
| acylglycerol catabolic process | 2 | 1.6 | 1.03 | -1.14 | -1.01 | 1.9 | -1.45 | -1.11 |
| membrane lipid catabolic process | 7 | -6.83 | -3.3 | -2.99 | 1.5 | -1.08 | -6.52 | -5.81 |
| membrane lipid biosynthetic process | 10 | -1.24 | 1.25 | 1.26 | 1.0 | 1.07 | -1.27 | 1.5 |
| phosphatidylcholine metabolic process | 2 | -1.14 | 1.06 | 1.34 | 1.11 | -1.05 | -1.18 | 1.93 |
| phosphatidylglycerol metabolic process | 1 | -3.36 | 1.4 | 1.32 | 1.25 | 1.35 | -2.25 | 1.21 |
| phosphatidic acid metabolic process | 1 | -3.17 | -1.24 | -2.56 | 1.51 | 2.33 | -10.59 | -5.09 |
| glycerophospholipid biosynthetic process | 29 | -1.18 | -1.03 | 1.13 | -1.01 | 1.18 | 1.02 | -1.13 |
| glycosylceramide catabolic process | 1 | -1.02 | 1.14 | 1.14 | 1.39 | 1.15 | 1.04 | -1.06 |
| glycosphingolipid catabolic process | 1 | -1.02 | 1.14 | 1.14 | 1.39 | 1.15 | 1.04 | -1.06 |
| heterocycle metabolic process | 248 | 1.2 | 1.03 | 1.01 | 1.0 | 1.02 | 1.19 | 1.01 |
| ether lipid metabolic process | 1 | 5.42 | -21.99 | -24.45 | -20.82 | -3.99 | -1.47 | 6.54 |
| glycerolipid metabolic process | 74 | 1.16 | -1.01 | 1.06 | -1.03 | 1.25 | 1.16 | 1.08 |
| glyoxylate metabolic process | 2 | 1.28 | -1.08 | -1.85 | -1.68 | -1.27 | 1.71 | -1.21 |
| phosphatidylinositol metabolic process | 56 | 1.09 | 1.02 | 1.12 | -1.01 | 1.23 | 1.24 | 1.03 |
| nicotinamide nucleotide metabolic process | 11 | 1.88 | 1.17 | -1.48 | -1.05 | 1.08 | 1.25 | 1.36 |
| nicotinate nucleotide metabolic process | 1 | 8.31 | 1.05 | -1.79 | -1.54 | -1.19 | 1.17 | 2.54 |
| S-adenosylmethionine metabolic process | 1 | 1.6 | 1.05 | 1.23 | -1.23 | 1.41 | 1.78 | 1.98 |
| protoporphyrinogen IX metabolic process | 1 | 1.62 | 1.26 | 1.09 | -1.07 | 1.01 | 1.21 | 1.32 |
| glycerolipid catabolic process | 2 | 1.6 | 1.03 | -1.14 | -1.01 | 1.9 | -1.45 | -1.11 |
| glycerol ether biosynthetic process | 4 | 3.17 | -2.08 | -1.12 | -2.44 | 1.05 | 1.32 | 2.23 |
| ceramide biosynthetic process | 2 | -1.99 | 1.43 | 1.45 | 1.05 | 1.2 | -1.4 | -1.17 |
| ceramide catabolic process | 3 | -3.87 | -1.6 | -1.06 | 1.31 | -1.04 | -6.06 | -3.71 |
| hypusine metabolic process | 2 | 1.15 | 1.07 | 1.04 | 1.07 | 1.11 | 1.08 | -1.05 |
| sphingoid metabolic process | 7 | -1.82 | -1.06 | 1.22 | 1.15 | 1.12 | -2.11 | -1.7 |
| sphingoid biosynthetic process | 2 | -1.99 | 1.43 | 1.45 | 1.05 | 1.2 | -1.4 | -1.17 |
| sphingoid catabolic process | 3 | -3.87 | -1.6 | -1.06 | 1.31 | -1.04 | -6.06 | -3.71 |
| xylosylprotein 4-beta-galactosyltransferase activity | 1 | 1.66 | 1.11 | 1.34 | 1.35 | 1.53 | 1.06 | -1.05 |
| glucosyltransferase activity | 13 | 1.53 | -1.16 | 1.01 | -1.3 | 1.09 | 1.41 | 1.06 |
| imaginal disc fusion, thorax closure | 15 | 1.03 | 1.24 | 1.46 | 1.13 | 1.3 | 1.26 | 1.21 |
| photoreceptor cell differentiation | 133 | 1.2 | 1.12 | 1.3 | 1.07 | 1.29 | 1.24 | 1.21 |
| regulation of photoreceptor cell differentiation | 27 | 1.02 | 1.11 | 1.32 | 1.05 | 1.3 | -1.01 | 1.16 |
| negative regulation of photoreceptor cell differentiation | 5 | -1.22 | 1.17 | 1.2 | 1.05 | 1.31 | -1.53 | 1.09 |
| positive regulation of photoreceptor cell differentiation | 8 | 1.3 | 1.17 | 1.47 | 1.09 | 1.39 | 1.19 | 1.25 |
| dosage compensation complex | 6 | 1.37 | 1.23 | 1.41 | 1.11 | 1.23 | 1.51 | 1.37 |
| development of primary female sexual characteristics | 17 | 1.27 | 1.11 | 1.08 | 1.01 | 1.21 | 1.15 | 1.13 |
| development of primary male sexual characteristics | 21 | 1.16 | 1.07 | 1.22 | 1.05 | 1.17 | -1.02 | 1.15 |
| photoreceptor cell fate commitment | 53 | 1.3 | 1.09 | 1.22 | 1.09 | 1.29 | 1.22 | 1.17 |
| regulation of Ras protein signal transduction | 76 | 1.02 | 1.17 | 1.37 | 1.05 | 1.32 | 1.3 | 1.27 |
| positive regulation of Ras protein signal transduction | 2 | 1.82 | 1.14 | 1.61 | 1.13 | 1.83 | 2.54 | 1.37 |
| negative regulation of Ras protein signal transduction | 7 | -1.35 | 1.4 | 1.44 | 1.09 | 1.53 | 1.07 | 1.44 |
| maintenance of pole plasm mRNA location | 3 | 1.53 | 1.04 | 1.01 | 1.11 | 1.01 | -1.06 | -1.06 |
| establishment of pole plasm mRNA localization | 1 | 1.48 | -1.03 | 1.16 | 1.42 | 1.58 | 1.23 | -1.33 |
| regulation of centriole replication | 2 | -1.05 | 1.36 | 1.16 | 1.21 | 1.29 | -1.05 | 1.21 |
| regulation of mitotic centrosome separation | 2 | -1.01 | 1.14 | 1.18 | 1.02 | 1.37 | 1.57 | 1.31 |
| regulation of centrosome cycle | 6 | 1.2 | 1.12 | 1.42 | 1.02 | 1.4 | 1.58 | 1.19 |
| regulation of organ growth | 37 | 1.36 | 1.2 | 1.4 | 1.1 | 1.41 | 1.43 | 1.25 |
| negative regulation of organ growth | 11 | 1.66 | 1.23 | 1.62 | 1.04 | 1.47 | 1.49 | 1.37 |
| positive regulation of organ growth | 11 | 1.2 | 1.28 | 1.51 | 1.16 | 1.65 | 1.9 | 1.4 |
| sphingolipid transporter activity | 1 | -2.39 | 1.47 | 1.51 | 1.07 | -1.25 | -2.16 | 1.24 |
| sphingolipid binding | 1 | -2.39 | 1.47 | 1.51 | 1.07 | -1.25 | -2.16 | 1.24 |
| regulation of insulin receptor signaling pathway | 10 | 1.25 | 1.09 | 1.37 | -1.04 | 1.17 | 1.16 | 1.59 |
| negative regulation of insulin receptor signaling pathway | 7 | 1.27 | 1.12 | 1.56 | -1.01 | 1.31 | 1.27 | 1.5 |
| positive regulation of insulin receptor signaling pathway | 2 | 1.0 | -1.0 | -1.17 | -1.4 | -1.23 | -1.17 | 2.26 |
| tetrahydrofolate metabolic process | 3 | 5.63 | -2.2 | -1.59 | -2.04 | -1.72 | 1.86 | 1.99 |
| tetrahydrofolate biosynthetic process | 1 | 33.5 | -5.63 | -5.79 | -6.03 | -4.58 | -1.32 | 6.94 |
| male sex differentiation | 23 | 1.21 | 1.08 | 1.19 | 1.05 | 1.15 | -1.03 | 1.14 |
| regulation of oviposition | 9 | -1.14 | -1.28 | -1.17 | -1.58 | 1.06 | -1.04 | 1.22 |
| dorsal closure, leading edge cell differentiation | 5 | 1.03 | 1.07 | 2.19 | -1.04 | 1.49 | -1.17 | 1.24 |
| dorsal closure, amnioserosa morphology change | 10 | 1.22 | 1.15 | 1.79 | 1.05 | 1.63 | 1.42 | 1.38 |
| amnioserosa maintenance | 5 | -1.1 | 1.14 | 1.38 | 1.06 | 1.23 | -1.08 | 1.88 |
| retinal cell programmed cell death | 11 | 1.07 | 1.14 | 1.21 | 1.16 | 1.27 | 1.33 | 1.16 |
| compound eye retinal cell programmed cell death | 10 | 1.07 | 1.14 | 1.21 | 1.15 | 1.3 | 1.4 | 1.22 |
| regulation of retinal cell programmed cell death | 17 | 1.06 | 1.05 | 1.39 | 1.1 | 1.32 | 1.05 | 1.22 |
| regulation of compound eye retinal cell programmed cell death | 16 | -1.01 | 1.05 | 1.48 | 1.08 | 1.37 | 1.0 | 1.24 |
| positive regulation of retinal cell programmed cell death | 7 | -1.3 | 1.11 | 1.24 | 1.18 | 1.41 | -1.33 | 1.3 |
| negative regulation of retinal cell programmed cell death | 7 | 1.19 | -1.06 | 1.77 | -1.01 | 1.35 | 1.16 | 1.21 |
| positive regulation of compound eye retinal cell programmed cell death | 7 | -1.3 | 1.11 | 1.24 | 1.18 | 1.41 | -1.33 | 1.3 |
| negative regulation of compound eye retinal cell programmed cell death | 7 | 1.19 | -1.06 | 1.77 | -1.01 | 1.35 | 1.16 | 1.21 |
| induction of retinal programmed cell death | 1 | 1.68 | -1.09 | 1.22 | 1.16 | 1.11 | 1.33 | 1.07 |
| induction of compound eye retinal cell programmed cell death | 1 | 1.68 | -1.09 | 1.22 | 1.16 | 1.11 | 1.33 | 1.07 |
| sperm competition | 15 | 1.28 | -1.27 | -1.33 | -1.33 | -1.2 | 1.37 | -1.28 |
| sperm storage | 8 | 1.66 | -1.63 | -1.61 | -1.67 | -1.3 | 1.96 | -1.5 |
| heterocycle catabolic process | 54 | 1.13 | -1.01 | 1.05 | 1.01 | 1.06 | 1.1 | 1.01 |
| insecticide catabolic process | 4 | -1.27 | -1.2 | -4.97 | -1.06 | -1.84 | -3.14 | 1.18 |
| galactoside 6-L-fucosyltransferase activity | 1 | 1.63 | 1.17 | -1.53 | -1.05 | -1.49 | 4.17 | 1.53 |
| muscle cell homeostasis | 8 | 1.75 | -1.12 | 1.33 | -1.09 | 1.16 | 1.67 | -1.03 |
| viral entry into host cell | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| microtubule polymerization | 2 | 1.26 | 1.08 | 1.08 | 1.42 | 1.42 | 1.11 | -1.04 |
| host cell surface binding | 1 | 2.63 | -1.41 | 1.69 | -1.89 | -1.48 | 14.61 | -1.02 |
| regulation of nucleocytoplasmic transport | 32 | 1.18 | 1.11 | 1.26 | 1.1 | 1.28 | 1.29 | 1.23 |
| negative regulation of nucleocytoplasmic transport | 6 | 2.03 | 1.13 | 1.29 | 1.08 | 1.28 | 1.86 | 1.59 |
| positive regulation of nucleocytoplasmic transport | 18 | 1.13 | 1.06 | 1.22 | 1.1 | 1.37 | 1.17 | 1.13 |
| regulation of protein export from nucleus | 1 | 1.39 | 1.22 | 1.43 | 1.19 | 1.67 | 1.14 | 1.14 |
| negative regulation of protein export from nucleus | 1 | 1.39 | 1.22 | 1.43 | 1.19 | 1.67 | 1.14 | 1.14 |
| lipid phosphorylation | 22 | 1.41 | 1.06 | 1.08 | -1.0 | 1.3 | 1.56 | 1.24 |
| glycolipid transport | 1 | -2.99 | 1.35 | -1.24 | 1.41 | -1.1 | -2.55 | -1.06 |
| phosphorylated carbohydrate dephosphorylation | 3 | 2.42 | 1.04 | 1.61 | -1.01 | 1.39 | 1.64 | 1.29 |
| phospholipid dephosphorylation | 4 | 1.17 | 1.2 | 1.42 | 1.09 | 1.23 | 1.3 | 1.22 |
| dorsal appendage formation | 31 | 1.38 | 1.07 | 1.32 | 1.03 | 1.3 | 1.21 | 1.3 |
| micropyle formation | 4 | -1.04 | 1.06 | 1.34 | 1.1 | 1.39 | -1.07 | 1.21 |
| branched duct epithelial cell fate determination, open tracheal system | 8 | 2.56 | -1.13 | -1.03 | -1.23 | 1.24 | 1.31 | 1.76 |
| filopodium assembly | 7 | 1.05 | 1.06 | 1.31 | 1.03 | 1.18 | 1.91 | 1.22 |
| phosphatidylinositol phosphorylation | 22 | 1.41 | 1.06 | 1.08 | -1.0 | 1.3 | 1.56 | 1.24 |
| inositol phosphate dephosphorylation | 3 | 2.42 | 1.04 | 1.61 | -1.01 | 1.39 | 1.64 | 1.29 |
| phosphatidylinositol dephosphorylation | 4 | 1.17 | 1.2 | 1.42 | 1.09 | 1.23 | 1.3 | 1.22 |
| isoprenoid transport | 1 | -6.48 | -6.48 | -8.02 | 2.81 | 1.04 | -8.53 | -8.65 |
| terpenoid transport | 1 | -6.48 | -6.48 | -8.02 | 2.81 | 1.04 | -8.53 | -8.65 |
| tetraterpenoid transport | 1 | -6.48 | -6.48 | -8.02 | 2.81 | 1.04 | -8.53 | -8.65 |
| carotenoid transport | 1 | -6.48 | -6.48 | -8.02 | 2.81 | 1.04 | -8.53 | -8.65 |
| regulation of hormone secretion | 2 | -1.01 | 1.27 | 1.27 | 1.12 | 1.08 | 1.42 | 1.16 |
| regulation of hormone biosynthetic process | 2 | -8.29 | -1.83 | 1.21 | 1.4 | 1.08 | -7.35 | -3.84 |
| regulation of lipid biosynthetic process | 6 | -3.09 | -1.16 | 1.01 | 1.27 | 1.37 | -3.44 | -2.07 |
| nucleoside triphosphate adenylate kinase activity | 1 | -1.71 | -1.23 | 1.26 | 1.63 | 1.63 | -3.14 | -2.07 |
| tetrahydrofolylpolyglutamate metabolic process | 1 | -2.47 | 1.42 | -1.59 | 1.58 | -1.34 | -2.64 | -1.65 |
| tetrahydrofolylpolyglutamate biosynthetic process | 1 | -2.47 | 1.42 | -1.59 | 1.58 | -1.34 | -2.64 | -1.65 |
| regulation of mitochondrial membrane permeability | 1 | 3.39 | -1.48 | 1.02 | -1.33 | 1.2 | 1.81 | 1.97 |
| tetrapyrrole binding | 134 | -1.06 | -1.16 | -1.25 | -1.06 | -1.02 | -1.06 | -1.12 |
| intracellular transport | 312 | 1.03 | 1.16 | 1.26 | 1.14 | 1.23 | 1.12 | 1.12 |
| ion homeostasis | 58 | 1.14 | 1.02 | 1.18 | -1.16 | -1.09 | 1.41 | 1.13 |
| transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer | 6 | 1.09 | -1.04 | -1.0 | -1.07 | -1.07 | 1.11 | -1.18 |
| transition metal ion binding | 1090 | 1.0 | 1.02 | 1.04 | 1.06 | 1.12 | 1.02 | -1.02 |
| transition metal ion transmembrane transporter activity | 19 | -1.09 | -1.13 | -1.04 | -1.14 | -1.01 | 1.11 | 1.1 |
| alpha-(1->3)-fucosyltransferase activity | 4 | -1.8 | 1.18 | 1.3 | 1.19 | 1.58 | -1.36 | -1.16 |
| alpha-(1->6)-fucosyltransferase activity | 1 | 1.63 | 1.17 | -1.53 | -1.05 | -1.49 | 4.17 | 1.53 |
| peptide-O-fucosyltransferase activity | 1 | -1.56 | -1.04 | -1.16 | 1.31 | -1.19 | -1.77 | -1.4 |
| ER retention sequence binding | 1 | -1.48 | 1.27 | 1.01 | 1.14 | -1.01 | -1.37 | 1.53 |
| regulation of neurotransmitter secretion | 11 | 1.29 | 1.05 | 1.09 | 1.01 | 1.04 | 1.26 | 1.14 |
| nucleotide phosphorylation | 8 | -1.1 | 1.06 | 1.21 | 1.26 | 1.33 | -1.22 | -1.25 |
| carboxylic acid transport | 54 | -1.28 | -1.1 | -1.53 | -1.18 | -1.03 | 1.07 | -1.12 |
| carboxylic acid transmembrane transporter activity | 86 | -1.42 | -1.03 | -1.43 | -1.14 | -1.01 | -1.05 | -1.13 |
| hydroxylysine metabolic process | 1 | -1.51 | -1.6 | -1.42 | 1.8 | -1.74 | 2.0 | -2.55 |
| hydroxylysine catabolic process | 1 | -1.51 | -1.6 | -1.42 | 1.8 | -1.74 | 2.0 | -2.55 |
| fatty-acyl-CoA biosynthetic process | 4 | 1.86 | 1.19 | 1.27 | 1.15 | 1.54 | 1.19 | 1.01 |
| cellular ketone body metabolic process | 1 | -1.42 | 1.61 | -1.39 | -1.13 | -1.97 | 3.73 | -1.22 |
| ketone body catabolic process | 1 | -1.42 | 1.61 | -1.39 | -1.13 | -1.97 | 3.73 | -1.22 |
| positive phototaxis | 2 | 1.78 | -1.02 | 1.5 | -1.1 | 1.34 | 2.13 | 1.12 |
| negative phototaxis | 1 | -1.56 | 1.9 | 2.35 | 1.42 | 1.33 | 1.03 | 1.8 |
| nonassociative learning | 7 | 1.25 | 1.0 | 1.45 | 1.14 | 1.48 | 1.86 | 1.46 |
| sensitization | 1 | 1.44 | -1.21 | -1.33 | 1.15 | 1.76 | 5.15 | 2.78 |
| 3'-phosphoadenosine 5'-phosphosulfate transport | 1 | 1.9 | 1.27 | 1.23 | 1.06 | 1.26 | 2.28 | 1.58 |
| 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity | 2 | 1.45 | 1.3 | 1.01 | 1.18 | 1.09 | 1.6 | 1.46 |
| histone acetyltransferase activity (H4-K16 specific) | 1 | 1.11 | 1.34 | 1.12 | 1.23 | 1.07 | 1.27 | 1.32 |
| histone methyltransferase activity (H3-K9 specific) | 4 | -1.11 | 1.13 | 1.21 | 1.02 | 1.08 | 1.06 | -1.09 |
| histone methyltransferase activity (H3-K36 specific) | 1 | 1.08 | 1.46 | 1.5 | -1.1 | 1.11 | 1.44 | 1.39 |
| histone methyltransferase activity (H3-K27 specific) | 3 | 1.02 | 1.31 | 1.33 | 1.16 | 1.21 | 1.01 | 1.04 |
| beta-1,4-mannosylglycolipid beta-1,3-N-acetylglucosaminyltransferase activity | 1 | 1.3 | 1.32 | 1.55 | 1.56 | 1.38 | -1.43 | -1.27 |
| protein heterodimerization activity | 59 | 1.16 | 1.07 | 1.25 | 1.01 | 1.07 | 1.14 | 1.08 |
| protein dimerization activity | 126 | 1.25 | 1.07 | 1.13 | -1.01 | 1.11 | 1.21 | 1.14 |
| N-acetyllactosamine beta-1,3-glucuronosyltransferase activity | 2 | -1.0 | -1.2 | -1.45 | -1.61 | -1.38 | 1.39 | 5.09 |
| asioloorosomucoid beta-1,3-glucuronosyltransferase activity | 2 | -1.0 | -1.2 | -1.45 | -1.61 | -1.38 | 1.39 | 5.09 |
| galactosyl beta-1,3 N-acetylgalactosamine beta-1,3-glucuronosyltransferase activity | 2 | -1.0 | -1.2 | -1.45 | -1.61 | -1.38 | 1.39 | 5.09 |
| 7-beta-hydroxysteroid dehydrogenase (NADP+) activity | 1 | 1.03 | -1.04 | 1.2 | -1.01 | 1.0 | 2.11 | -1.07 |
| testosterone dehydrogenase (NAD+) activity | 1 | 1.03 | -1.04 | 1.2 | -1.01 | 1.0 | 2.11 | -1.07 |
| vitamin-K-epoxide reductase (warfarin-sensitive) activity | 1 | 1.07 | 1.03 | -1.07 | 1.05 | -1.1 | -1.02 | -1.18 |
| inositol tetrakisphosphate 5-kinase activity | 1 | -1.15 | 1.56 | -1.07 | 1.07 | -1.15 | -1.25 | 1.09 |
| glycosylphosphatidylinositol diacylglycerol-lyase activity | 2 | 2.29 | 1.0 | -1.19 | -1.05 | 1.0 | 1.66 | 1.76 |
| nucleoside-triphosphate diphosphatase activity | 1 | -1.36 | -1.27 | 1.07 | 1.7 | 1.82 | -3.47 | -2.13 |
| N-acylneuraminate-9-phosphate synthase activity | 1 | -1.06 | 1.35 | 1.0 | -1.13 | -1.03 | 1.09 | -1.01 |
| regulation of response to osmotic stress | 1 | -6.38 | -3.77 | -11.1 | -5.2 | -1.19 | 28.07 | -1.66 |
| vesicle transport along microtubule | 1 | 1.21 | 1.12 | 1.66 | 1.2 | 1.79 | 2.51 | 1.99 |
| mitochondrion transport along microtubule | 5 | -1.72 | 1.29 | 1.54 | 1.18 | 1.39 | 1.16 | 1.27 |
| 3',5'-cyclic-GMP phosphodiesterase activity | 5 | 1.32 | 1.17 | 1.39 | 1.05 | 1.28 | 2.17 | 1.4 |
| acyl-CoA hydrolase activity | 2 | 1.13 | 1.03 | -1.08 | 1.05 | 1.02 | 1.1 | -1.09 |
| alanine-oxo-acid transaminase activity | 1 | -2.33 | 1.13 | 1.14 | 1.09 | 1.76 | 1.77 | -1.09 |
| bis(5'-adenosyl)-triphosphatase activity | 1 | 2.29 | 1.26 | 1.38 | 1.23 | 1.51 | 1.35 | -1.03 |
| cysteamine dioxygenase activity | 1 | 2.58 | 1.06 | 1.36 | 1.25 | 1.89 | 1.41 | 1.2 |
| ecdysone oxidase activity | 1 | 1.11 | 1.03 | -1.05 | 1.12 | 4.21 | 1.26 | -1.02 |
| insulin-like growth factor receptor signaling pathway | 2 | -5.92 | 1.37 | 1.08 | 1.24 | 1.25 | -4.11 | -1.32 |
| vascular endothelial growth factor receptor signaling pathway | 4 | -1.2 | 1.05 | 1.06 | 1.13 | 1.34 | -1.18 | 1.04 |
| receptor antagonist activity | 2 | 1.75 | 1.03 | 1.01 | 1.06 | 1.08 | 1.1 | 1.1 |
| regulation of melanin biosynthetic process | 1 | 1.36 | 1.09 | 1.02 | -1.03 | -1.16 | 1.04 | 1.01 |
| negative regulation of melanin biosynthetic process | 1 | 1.36 | 1.09 | 1.02 | -1.03 | -1.16 | 1.04 | 1.01 |
| regulation of nuclear mRNA splicing, via spliceosome | 58 | 1.19 | 1.18 | 1.28 | 1.17 | 1.24 | 1.4 | 1.08 |
| negative regulation of nuclear mRNA splicing, via spliceosome | 3 | 1.33 | 1.28 | 1.25 | 1.28 | 1.23 | 1.34 | 1.03 |
| mRNA 5'-UTR binding | 1 | -1.51 | 1.63 | 1.29 | 1.08 | 1.13 | 1.32 | 1.66 |
| monosaccharide binding | 18 | -1.33 | -1.12 | 1.17 | -1.04 | -1.09 | -1.32 | 1.07 |
| heme o metabolic process | 1 | -1.18 | 1.24 | 1.2 | 1.18 | -1.04 | 1.79 | -1.68 |
| heme O biosynthetic process | 1 | -1.18 | 1.24 | 1.2 | 1.18 | -1.04 | 1.79 | -1.68 |
| central complex development | 6 | -1.05 | 1.15 | 1.33 | 1.1 | 1.3 | 1.02 | 1.11 |
| cofactor binding | 179 | 1.0 | 1.05 | -1.16 | 1.01 | 1.02 | -1.02 | 1.0 |
| quinone binding | 6 | 1.1 | 1.03 | 1.2 | 1.04 | 1.1 | 1.42 | -1.29 |
| ubiquinone binding | 1 | 1.51 | -1.05 | 1.4 | 1.02 | 1.33 | 1.88 | -1.23 |
| focal adhesion assembly | 1 | -1.1 | -1.15 | -1.07 | -1.18 | 1.52 | 1.28 | 1.24 |
| regulation of post-mating oviposition | 5 | -1.35 | -1.04 | 1.16 | -1.45 | 1.28 | -1.43 | 1.45 |
| mating behavior, sex discrimination | 3 | -1.02 | 1.04 | -1.06 | -1.01 | -1.08 | -1.1 | 1.0 |
| R1/R6 cell differentiation | 5 | 1.29 | 1.11 | 1.18 | -1.01 | 1.23 | 1.16 | 1.34 |
| R1/R6 development | 1 | 2.43 | 1.22 | 1.86 | 1.12 | 2.17 | 2.26 | 2.36 |
| R2/R5 cell differentiation | 1 | 1.01 | 1.09 | 1.11 | -1.09 | -1.01 | 1.02 | -1.12 |
| R3/R4 cell differentiation | 15 | 1.5 | 1.09 | 1.17 | 1.02 | 1.41 | 1.3 | 1.28 |
| R3/R4 development | 1 | -1.12 | -1.06 | -1.22 | 1.01 | -1.04 | -1.1 | 1.01 |
| compound eye corneal lens development | 2 | 2.59 | 1.11 | 1.99 | 1.1 | 2.05 | 1.65 | 1.78 |
| negative gravitaxis | 4 | 1.28 | -1.05 | 1.11 | 1.0 | 1.1 | 1.42 | 1.06 |
| male courtship behavior, veined wing extension | 14 | 1.06 | 1.04 | 1.41 | 1.13 | 1.09 | -1.02 | -1.01 |
| developmental pigmentation | 55 | 1.19 | -1.1 | -1.11 | 1.06 | -1.01 | -1.08 | -1.01 |
| cuticle pigmentation | 11 | 1.56 | -1.44 | -1.19 | -1.25 | -1.22 | 1.2 | 1.58 |
| regulation of developmental pigmentation | 21 | -1.11 | 1.02 | 1.05 | 1.01 | 1.13 | -1.14 | 1.25 |
| sex-specific pigmentation | 3 | 1.97 | 1.09 | 1.06 | -1.03 | 1.05 | 1.06 | 1.01 |
| compound eye pigmentation | 12 | 1.67 | -1.11 | -1.16 | -1.06 | -1.06 | -1.02 | 1.25 |
| regulation of eye pigmentation | 5 | -1.48 | 1.1 | 1.53 | 1.11 | 1.49 | -1.35 | 1.53 |
| regulation of compound eye pigmentation | 5 | -1.48 | 1.1 | 1.53 | 1.11 | 1.49 | -1.35 | 1.53 |
| regulation of cuticle pigmentation | 13 | -1.19 | -1.03 | -1.1 | -1.02 | 1.03 | -1.11 | 1.21 |
| positive regulation of cuticle pigmentation | 1 | 1.47 | 1.1 | -1.04 | -1.06 | 1.06 | -1.18 | 1.3 |
| regulation of adult chitin-containing cuticle pigmentation | 12 | -1.25 | -1.04 | -1.11 | -1.02 | 1.03 | -1.11 | 1.2 |
| adult chitin-containing cuticle pigmentation | 5 | 1.15 | -1.36 | -1.08 | -1.2 | -1.43 | 1.07 | 1.86 |
| negative regulation of developmental pigmentation | 2 | 2.98 | 1.16 | 1.17 | 1.04 | 1.29 | 1.15 | 1.23 |
| positive regulation of developmental pigmentation | 2 | 1.12 | 1.03 | -1.09 | -1.11 | -1.16 | -1.13 | -1.07 |
| regulation of male pigmentation | 1 | 1.95 | -1.01 | 1.39 | -1.03 | 1.33 | 1.49 | 1.42 |
| negative regulation of male pigmentation | 1 | 1.95 | -1.01 | 1.39 | -1.03 | 1.33 | 1.49 | 1.42 |
| male pigmentation | 1 | -1.17 | -1.03 | -1.15 | -1.16 | -1.43 | -1.09 | -1.47 |
| antennal joint development | 4 | -1.02 | 1.09 | 1.04 | 1.04 | -1.02 | 1.19 | -1.03 |
| anterior/posterior lineage restriction, imaginal disc | 4 | 1.14 | -1.2 | 1.12 | 1.11 | 1.19 | -1.1 | -1.24 |
| wing disc anterior/posterior pattern formation | 12 | 1.55 | -1.26 | -1.12 | -1.07 | 1.12 | 1.32 | -1.04 |
| calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity | 1 | -1.09 | -1.0 | 1.07 | 1.3 | -1.04 | -1.15 | -1.07 |
| somatic stem cell division | 38 | -1.22 | 1.15 | 1.34 | 1.17 | 1.41 | -1.02 | 1.03 |
| establishment of body hair or bristle planar orientation | 5 | 2.56 | -1.0 | 1.21 | -1.16 | 1.38 | 2.42 | 1.68 |
| establishment of body hair planar orientation | 1 | 1.7 | -1.08 | -1.05 | -1.16 | 1.03 | 1.04 | 1.05 |
| establishment of thoracic bristle planar orientation | 2 | 1.54 | 1.4 | 2.06 | 1.24 | 1.94 | 2.07 | 1.64 |
| female germ-line stem cell division | 10 | 1.1 | 1.17 | 1.12 | 1.12 | 1.15 | 1.0 | 1.13 |
| male germ-line stem cell division | 5 | 1.44 | 1.28 | 1.52 | 1.25 | 1.58 | 1.33 | 1.36 |
| germ-line cyst formation | 30 | 1.15 | 1.12 | 1.06 | 1.13 | 1.31 | 1.13 | 1.08 |
| female germ-line cyst formation | 11 | 1.0 | 1.02 | 1.1 | 1.01 | 1.25 | 1.47 | 1.16 |
| male germ-line cyst formation | 16 | 1.24 | 1.16 | 1.05 | 1.27 | 1.48 | -1.14 | -1.0 |
| spermatocyte division | 12 | 1.41 | 1.22 | 1.06 | 1.3 | 1.57 | -1.11 | 1.0 |
| germ-line cyst encapsulation | 10 | 1.3 | 1.19 | 1.37 | 1.1 | 1.51 | 1.67 | 1.42 |
| female germ-line cyst encapsulation | 10 | 1.3 | 1.19 | 1.37 | 1.1 | 1.51 | 1.67 | 1.42 |
| male germ-line cyst encapsulation | 1 | 2.54 | -1.11 | -1.09 | 1.14 | 1.59 | 3.23 | 1.89 |
| germarium-derived cystoblast division | 2 | -1.06 | 1.04 | 1.19 | 1.36 | 1.87 | -1.4 | -1.21 |
| regulation of synaptic plasticity | 12 | -1.02 | 1.07 | 1.27 | 1.06 | 1.18 | 1.14 | 1.24 |
| regulation of neuronal synaptic plasticity | 7 | 1.02 | 1.03 | 1.32 | 1.05 | 1.12 | 1.11 | 1.29 |
| regulation of long-term neuronal synaptic plasticity | 1 | -1.14 | 1.2 | 1.7 | 1.09 | 2.04 | 1.39 | 1.74 |
| regulation of short-term neuronal synaptic plasticity | 5 | -1.17 | 1.04 | 1.36 | 1.07 | 1.04 | 1.06 | 1.15 |
| activin receptor complex | 2 | -1.14 | 1.57 | 1.93 | 1.23 | 1.67 | 1.43 | 1.4 |
| activin binding | 2 | -1.14 | 1.57 | 1.93 | 1.23 | 1.67 | 1.43 | 1.4 |
| Set1C/COMPASS complex | 8 | 1.45 | 1.14 | 1.33 | 1.24 | 1.41 | 1.36 | 1.13 |
| wing disc dorsal/ventral pattern formation | 40 | 1.21 | 1.03 | 1.1 | 1.01 | 1.29 | 1.34 | 1.31 |
| Golgi vesicle transport | 49 | -1.1 | 1.16 | 1.21 | 1.13 | 1.21 | 1.13 | 1.34 |
| Golgi vesicle budding | 1 | -2.28 | 1.35 | -1.05 | 1.06 | -1.19 | -1.24 | 1.52 |
| vesicle targeting, to, from or within Golgi | 1 | -1.49 | 1.52 | 1.25 | 1.12 | 1.19 | 1.46 | 1.78 |
| vesicle targeting, rough ER to cis-Golgi | 1 | -1.49 | 1.52 | 1.25 | 1.12 | 1.19 | 1.46 | 1.78 |
| COPII vesicle coating | 1 | -1.49 | 1.52 | 1.25 | 1.12 | 1.19 | 1.46 | 1.78 |
| Golgi vesicle fusion to target membrane | 1 | -1.26 | 1.22 | 1.43 | 1.12 | 1.39 | 1.17 | -1.02 |
| male gamete generation | 151 | 1.07 | 1.14 | 1.22 | 1.07 | 1.24 | 1.12 | 1.11 |
| mitochondrial iron ion transport | 1 | -1.62 | 1.12 | -1.09 | 1.38 | 1.65 | -1.57 | 1.22 |
| lauric acid metabolic process | 1 | 1.6 | 1.35 | -1.72 | -1.6 | -1.17 | -1.33 | 1.5 |
| flap endonuclease activity | 1 | 2.54 | 1.11 | 1.06 | 1.23 | 1.48 | 1.16 | -1.04 |
| regulation of receptor-mediated endocytosis | 6 | 1.03 | -1.04 | 1.28 | 1.02 | 1.13 | 1.2 | 1.06 |
| positive regulation of receptor-mediated endocytosis | 5 | 1.04 | -1.02 | 1.26 | 1.02 | 1.14 | 1.19 | 1.06 |
| negative regulation of receptor-mediated endocytosis | 1 | -1.04 | -1.14 | 1.37 | -1.01 | 1.09 | 1.23 | 1.09 |
| determination of dorsal/ventral asymmetry | 1 | -1.43 | -1.07 | -1.23 | -1.08 | -1.37 | -1.24 | 1.3 |
| determination of ventral identity | 1 | -1.43 | -1.07 | -1.23 | -1.08 | -1.37 | -1.24 | 1.3 |
| response to pain | 2 | 1.88 | -1.18 | 1.22 | 1.16 | 1.43 | 1.98 | 1.19 |
| behavioral response to pain | 2 | 1.88 | -1.18 | 1.22 | 1.16 | 1.43 | 1.98 | 1.19 |
| clathrin coat assembly | 1 | -1.06 | 1.07 | 1.15 | -1.0 | 1.23 | 1.29 | 1.44 |
| vesicle docking | 27 | -1.1 | 1.16 | 1.4 | 1.15 | 1.26 | 1.14 | 1.1 |
| vesicle fusion with Golgi apparatus | 1 | -1.71 | 1.25 | 1.03 | 1.37 | 1.03 | -1.21 | 1.5 |
| organelle fusion | 22 | -1.09 | 1.09 | 1.26 | 1.2 | 1.3 | 1.01 | 1.01 |
| organelle fission | 154 | 1.25 | 1.09 | 1.26 | 1.12 | 1.32 | 1.24 | 1.11 |
| calcium-dependent protein binding | 1 | -1.16 | -1.03 | -1.2 | -1.35 | 1.14 | -1.16 | 1.14 |
| mitochondrion distribution | 4 | -1.16 | 1.47 | 1.48 | 1.13 | 1.3 | 1.01 | 1.31 |
| intracellular distribution of mitochondria | 3 | -1.04 | 1.59 | 1.47 | 1.15 | 1.39 | 1.02 | 1.35 |
| mesoderm morphogenesis | 27 | 1.38 | 1.0 | 1.14 | -1.07 | 1.1 | 1.58 | 1.18 |
| mesodermal cell differentiation | 16 | 1.47 | 1.02 | 1.02 | -1.04 | 1.04 | 1.32 | 1.13 |
| retinoic acid receptor signaling pathway | 1 | 1.03 | 1.25 | 1.07 | 1.02 | 1.08 | 1.24 | 1.08 |
| endosomal lumen acidification | 2 | 1.23 | 1.13 | 1.3 | 1.15 | 1.45 | 1.18 | 1.47 |
| cell development | 815 | 1.17 | 1.08 | 1.24 | 1.04 | 1.25 | 1.26 | 1.2 |
| cell maturation | 116 | 1.16 | 1.11 | 1.31 | 1.06 | 1.29 | 1.43 | 1.21 |
| perinuclear region of cytoplasm | 31 | -1.12 | 1.12 | 1.34 | 1.08 | 1.32 | 1.38 | 1.19 |
| coated membrane | 25 | -1.02 | 1.18 | 1.24 | 1.13 | 1.15 | 1.19 | 1.33 |
| oogenesis | 421 | 1.13 | 1.11 | 1.26 | 1.07 | 1.26 | 1.21 | 1.2 |
| replication fork protection | 1 | -1.49 | 1.01 | -1.08 | 1.09 | -1.02 | -1.34 | -1.39 |
| synaptic vesicle endocytosis | 44 | 1.07 | 1.17 | 1.41 | 1.1 | 1.29 | 1.39 | 1.22 |
| synaptic vesicle transport | 94 | 1.0 | 1.11 | 1.34 | 1.11 | 1.24 | 1.22 | 1.14 |
| anterograde synaptic vesicle transport | 3 | -1.11 | 1.21 | 1.44 | 1.19 | 1.33 | 1.23 | 1.39 |
| retrograde synaptic vesicle transport | 1 | -1.08 | 1.01 | 1.18 | 1.12 | 1.07 | -1.04 | 1.06 |
| synaptic vesicle membrane organization | 1 | -1.53 | 1.15 | 2.49 | 1.01 | 1.71 | 1.37 | 1.07 |
| signal recognition particle | 9 | -1.25 | 1.37 | 1.01 | 1.14 | -1.09 | -1.57 | 1.26 |
| regulation of timing of cell differentiation | 1 | -1.09 | 1.32 | 1.47 | 1.06 | 1.16 | 1.21 | 1.23 |
| rhythmic process | 57 | 1.07 | -1.04 | 1.05 | -1.19 | -1.1 | 1.29 | 1.15 |
| circadian behavior | 44 | -1.03 | 1.02 | 1.17 | -1.13 | -1.04 | 1.31 | 1.08 |
| organ development | 955 | 1.15 | 1.06 | 1.2 | 1.03 | 1.21 | 1.23 | 1.17 |
| spermatid differentiation | 82 | 1.02 | 1.13 | 1.3 | 1.04 | 1.31 | 1.2 | 1.17 |
| CARD domain binding | 1 | 1.78 | 1.11 | 1.56 | 1.24 | 1.76 | -1.03 | 1.11 |
| positive regulation of biological process | 520 | 1.13 | 1.08 | 1.2 | 1.04 | 1.22 | 1.19 | 1.19 |
| negative regulation of biological process | 679 | 1.2 | 1.08 | 1.24 | 1.06 | 1.24 | 1.26 | 1.16 |
| positive regulation of behavior | 2 | 1.24 | 1.16 | 1.04 | -1.02 | 1.05 | 1.25 | 1.29 |
| negative regulation of behavior | 1 | 1.49 | 1.07 | 1.15 | 1.04 | -1.03 | 1.53 | -1.09 |
| positive regulation of cellular process | 486 | 1.12 | 1.08 | 1.2 | 1.04 | 1.22 | 1.19 | 1.2 |
| negative regulation of cellular process | 586 | 1.18 | 1.09 | 1.25 | 1.06 | 1.25 | 1.26 | 1.18 |
| negative regulation of viral reproduction | 4 | 1.27 | 1.23 | 1.52 | 1.17 | 1.6 | 1.47 | 1.34 |
| imaginal disc-derived wing expansion | 6 | 2.01 | -1.04 | 1.17 | -1.02 | 1.2 | 1.5 | 1.51 |
| beta-1,3-galactosyltransferase activity | 10 | 1.35 | 1.03 | 1.08 | 1.01 | -1.01 | 1.23 | 1.16 |
| hemopoietic or lymphoid organ development | 52 | 1.16 | 1.07 | 1.33 | 1.05 | 1.3 | 1.2 | 1.34 |
| response to steroid hormone stimulus | 41 | 1.14 | 1.06 | 1.3 | 1.01 | 1.23 | 1.41 | 1.22 |
| digestive tract morphogenesis | 61 | 1.11 | -1.08 | 1.05 | -1.02 | 1.31 | 1.24 | 1.17 |
| metalloenzyme inhibitor activity | 1 | 8.96 | -2.17 | 1.12 | -1.71 | -1.16 | 3.1 | 2.84 |
| post-embryonic organ morphogenesis | 285 | 1.23 | 1.07 | 1.25 | 1.02 | 1.3 | 1.3 | 1.29 |
| digestive tract development | 87 | -1.01 | -1.06 | 1.09 | -1.05 | 1.19 | 1.11 | 1.14 |
| embryonic digestive tract development | 1 | -1.17 | -1.0 | -1.0 | -1.0 | -1.17 | -1.16 | 1.21 |
| ectodermal digestive tract morphogenesis | 4 | 1.19 | 1.28 | -1.02 | 1.12 | 1.64 | 1.2 | 1.13 |
| embryonic organ development | 24 | -1.01 | 1.08 | 1.33 | 1.05 | 1.28 | 1.06 | 1.27 |
| post-embryonic organ development | 300 | 1.23 | 1.07 | 1.25 | 1.02 | 1.29 | 1.29 | 1.29 |
| regulation of post-embryonic development | 5 | 1.14 | 1.16 | -1.03 | -1.07 | -1.0 | 1.5 | 1.12 |
| regulation of response to stimulus | 449 | 1.14 | 1.09 | 1.24 | 1.03 | 1.21 | 1.24 | 1.19 |
| positive regulation of response to stimulus | 91 | 1.19 | 1.1 | 1.22 | 1.05 | 1.25 | 1.21 | 1.24 |
| negative regulation of response to stimulus | 151 | 1.23 | 1.1 | 1.3 | 1.05 | 1.27 | 1.31 | 1.24 |
| developmental cell growth | 36 | 1.21 | 1.12 | 1.29 | 1.01 | 1.33 | 1.3 | 1.21 |
| developmental growth | 107 | 1.31 | 1.12 | 1.33 | 1.03 | 1.34 | 1.41 | 1.29 |
| eye morphogenesis | 209 | 1.17 | 1.09 | 1.26 | 1.05 | 1.25 | 1.21 | 1.26 |
| embryonic morphogenesis | 201 | 1.18 | 1.04 | 1.26 | 1.01 | 1.29 | 1.31 | 1.22 |
| oocyte development | 102 | 1.15 | 1.14 | 1.34 | 1.08 | 1.28 | 1.39 | 1.24 |
| oocyte morphogenesis | 1 | 1.25 | 1.21 | 2.24 | 1.64 | 2.85 | 9.21 | 1.2 |
| reproductive structure development | 42 | 1.01 | 1.04 | 1.05 | 1.07 | 1.24 | -1.01 | 1.0 |
| multicellular organismal reproductive process | 651 | 1.12 | 1.07 | 1.19 | 1.04 | 1.21 | 1.19 | 1.17 |
| cellular process involved in reproduction | 549 | 1.13 | 1.09 | 1.21 | 1.05 | 1.26 | 1.19 | 1.17 |
| embryonic hindgut morphogenesis | 45 | 1.28 | -1.09 | 1.1 | -1.04 | 1.28 | 1.38 | 1.09 |
| myoblast cell fate commitment | 1 | -1.07 | -1.09 | 1.09 | -1.1 | 1.09 | -1.05 | 1.02 |
| regulation of muscle organ development | 54 | 1.15 | 1.07 | 1.22 | 1.02 | 1.13 | 1.16 | 1.15 |
| negative regulation of muscle organ development | 3 | 1.43 | 1.06 | 1.43 | -1.11 | 1.0 | 1.31 | -1.06 |
| positive regulation of muscle organ development | 6 | 2.17 | -1.15 | 1.0 | -1.2 | 1.03 | 1.63 | 1.19 |
| regulation of developmental growth | 66 | 1.12 | 1.14 | 1.34 | 1.08 | 1.24 | 1.16 | 1.2 |
| positive regulation of developmental growth | 12 | -1.03 | 1.16 | 1.18 | 1.12 | 1.31 | 1.14 | 1.28 |
| negative regulation of developmental growth | 18 | -1.04 | 1.11 | 1.34 | 1.12 | 1.2 | -1.08 | 1.05 |
| regulation of skeletal muscle tissue development | 39 | -1.02 | 1.12 | 1.32 | 1.09 | 1.24 | 1.09 | 1.21 |
| muscle organ morphogenesis | 4 | 2.88 | -1.16 | 1.03 | -1.17 | 1.2 | 1.86 | 1.03 |
| organ formation | 41 | 1.13 | 1.03 | 1.17 | 1.07 | 1.3 | 1.14 | 1.19 |
| neuron fate commitment | 58 | 1.29 | 1.09 | 1.23 | 1.1 | 1.29 | 1.22 | 1.17 |
| neuron fate specification | 12 | 1.09 | 1.06 | 1.08 | 1.18 | 1.13 | -1.18 | -1.07 |
| neuron development | 401 | 1.13 | 1.09 | 1.25 | 1.03 | 1.27 | 1.26 | 1.25 |
| cell morphogenesis involved in neuron differentiation | 329 | 1.14 | 1.09 | 1.26 | 1.04 | 1.27 | 1.25 | 1.26 |
| collateral sprouting | 3 | -1.12 | 1.14 | 1.65 | 1.16 | 1.31 | -1.01 | 1.22 |
| collateral sprouting of injured axon | 3 | -1.12 | 1.14 | 1.65 | 1.16 | 1.31 | -1.01 | 1.22 |
| axon extension | 21 | 1.21 | 1.14 | 1.27 | 1.01 | 1.26 | 1.1 | 1.2 |
| axon extension involved in development | 6 | 1.15 | 1.12 | 1.61 | 1.08 | 1.39 | 1.07 | 1.23 |
| response to axon injury | 4 | -1.08 | 1.15 | 1.5 | 1.12 | 1.22 | -1.04 | 1.21 |
| sprouting of injured axon | 3 | -1.12 | 1.14 | 1.65 | 1.16 | 1.31 | -1.01 | 1.22 |
| generation of neurons | 540 | 1.12 | 1.09 | 1.26 | 1.04 | 1.27 | 1.24 | 1.22 |
| instar larval or pupal morphogenesis | 353 | 1.18 | 1.06 | 1.22 | 1.03 | 1.28 | 1.26 | 1.24 |
| proboscis development | 6 | 1.03 | 1.0 | 1.19 | 1.01 | 1.08 | 1.14 | 1.54 |
| tissue morphogenesis | 259 | 1.2 | 1.04 | 1.27 | 1.02 | 1.28 | 1.32 | 1.25 |
| epidermis morphogenesis | 5 | 1.0 | 1.15 | 1.73 | 1.35 | 1.48 | 1.06 | 1.17 |
| system development | 1724 | 1.15 | 1.05 | 1.18 | 1.04 | 1.2 | 1.21 | 1.14 |
| gland development | 163 | 1.1 | 1.01 | 1.16 | 1.05 | 1.25 | 1.17 | 1.18 |
| proboscis morphogenesis | 1 | -1.31 | 1.06 | 2.72 | 1.23 | 2.11 | 1.21 | 1.47 |
| haltere morphogenesis | 1 | 5.22 | -1.53 | 1.19 | 1.44 | -1.43 | -1.39 | -1.19 |
| appendage development | 238 | 1.25 | 1.06 | 1.22 | 1.01 | 1.29 | 1.33 | 1.3 |
| imaginal disc-derived appendage development | 236 | 1.25 | 1.06 | 1.22 | 1.01 | 1.29 | 1.33 | 1.3 |
| cardiac muscle tissue development | 5 | 1.62 | -1.11 | 1.15 | -1.06 | 1.18 | 1.57 | 1.06 |
| skeletal muscle fiber development | 47 | 1.2 | 1.17 | 1.29 | 1.08 | 1.21 | 1.33 | 1.23 |
| regulation of skeletal muscle fiber development | 39 | -1.02 | 1.12 | 1.32 | 1.09 | 1.24 | 1.09 | 1.21 |
| muscle fiber development | 55 | 1.21 | 1.17 | 1.27 | 1.04 | 1.19 | 1.31 | 1.24 |
| compound eye development | 252 | 1.14 | 1.1 | 1.26 | 1.07 | 1.26 | 1.21 | 1.24 |
| compound eye corneal lens morphogenesis | 1 | 2.43 | 1.22 | 1.86 | 1.12 | 2.17 | 2.26 | 2.36 |
| pigment granule organization | 11 | 1.07 | 1.19 | 1.37 | 1.13 | 1.22 | 1.15 | 1.17 |
| branching morphogenesis of a tube | 45 | 1.12 | 1.12 | 1.47 | 1.03 | 1.34 | 1.29 | 1.3 |
| mesenchymal cell differentiation | 1 | 1.47 | 1.13 | -1.02 | 1.1 | -1.07 | -1.03 | 1.1 |
| calcium-induced calcium release activity | 1 | 2.63 | 1.01 | 1.53 | -1.05 | 1.46 | 2.52 | 1.76 |
| presynaptic active zone | 10 | 1.01 | 1.21 | 1.32 | 1.11 | 1.33 | 1.13 | 1.22 |
| presynaptic cytoskeletal matrix assembled at active zones | 1 | 1.09 | 1.05 | 1.11 | -1.02 | 1.12 | -1.14 | -1.07 |
| cytoskeletal matrix organization at active zone | 3 | 1.12 | 1.05 | 1.11 | 1.12 | 1.08 | -1.03 | -1.02 |
| maintenance of presynaptic active zone structure | 6 | -1.17 | 1.19 | 1.5 | 1.02 | 1.42 | 1.14 | 1.22 |
| calcium ion-dependent exocytosis of neurotransmitter | 1 | 1.37 | 1.07 | 1.15 | 1.12 | 1.04 | 1.3 | 1.03 |
| antennal morphogenesis | 13 | -1.02 | 1.09 | 1.27 | 1.06 | 1.16 | 1.4 | 1.12 |
| antennal joint morphogenesis | 1 | 1.21 | 1.22 | 1.16 | 1.1 | 1.2 | 1.57 | 1.03 |
| notum morphogenesis | 4 | -1.53 | 1.03 | 1.15 | 1.08 | 1.29 | -1.65 | -1.02 |
| imaginal disc-derived male genitalia morphogenesis | 13 | 1.4 | 1.09 | 1.4 | 1.06 | 1.26 | 1.05 | 1.3 |
| imaginal disc-derived female genitalia morphogenesis | 1 | 1.36 | 1.32 | 2.03 | 1.27 | 1.67 | 1.42 | 1.61 |
| imaginal disc-derived genitalia morphogenesis | 13 | 1.4 | 1.09 | 1.4 | 1.06 | 1.26 | 1.05 | 1.3 |
| genitalia development | 25 | 1.49 | 1.05 | 1.16 | 1.03 | 1.12 | 1.12 | 1.21 |
| female genitalia morphogenesis | 1 | 1.36 | 1.32 | 2.03 | 1.27 | 1.67 | 1.42 | 1.61 |
| male genitalia morphogenesis | 14 | 1.37 | 1.09 | 1.36 | 1.06 | 1.24 | 1.04 | 1.28 |
| neuron projection morphogenesis | 325 | 1.14 | 1.09 | 1.25 | 1.03 | 1.26 | 1.25 | 1.26 |
| dendrite morphogenesis | 132 | 1.18 | 1.12 | 1.28 | 1.11 | 1.28 | 1.22 | 1.16 |
| regulation of dendrite morphogenesis | 19 | 1.2 | 1.13 | 1.22 | 1.12 | 1.33 | 1.36 | 1.19 |
| ocellus morphogenesis | 1 | 1.15 | 1.0 | -1.08 | 1.01 | -1.06 | -1.04 | 1.1 |
| regulation of axon extension involved in axon guidance | 2 | -1.17 | 1.18 | 1.16 | 1.13 | 1.08 | -1.15 | -1.0 |
| axon extension involved in axon guidance | 2 | -1.19 | -1.04 | -1.25 | -1.1 | -1.33 | -1.25 | -1.21 |
| brain morphogenesis | 15 | 1.06 | 1.14 | 1.38 | 1.07 | 1.42 | 1.16 | 1.21 |
| anatomical structure development | 2169 | 1.15 | 1.05 | 1.16 | 1.04 | 1.18 | 1.2 | 1.13 |
| cell projection morphogenesis | 358 | 1.11 | 1.08 | 1.23 | 1.04 | 1.24 | 1.23 | 1.23 |
| formation of anatomical boundary | 40 | 1.1 | -1.02 | 1.14 | -1.0 | 1.24 | 1.07 | 1.39 |
| stem cell differentiation | 55 | 1.1 | 1.12 | 1.3 | 1.12 | 1.35 | 1.1 | 1.2 |
| stem cell development | 50 | 1.1 | 1.12 | 1.31 | 1.12 | 1.35 | 1.12 | 1.22 |
| stem cell fate commitment | 5 | 1.31 | 1.03 | 1.18 | 1.08 | 1.47 | 1.16 | 1.29 |
| cellular developmental process | 1656 | 1.14 | 1.07 | 1.19 | 1.06 | 1.2 | 1.19 | 1.12 |
| cell motility | 198 | 1.13 | 1.09 | 1.32 | 1.04 | 1.35 | 1.22 | 1.2 |
| multicellular organismal homeostasis | 22 | 1.28 | -1.0 | 1.13 | -1.11 | 1.22 | 1.43 | 1.52 |
| chemical homeostasis | 84 | -1.06 | -1.02 | 1.05 | -1.1 | -1.06 | 1.13 | 1.03 |
| peripheral nervous system neuron differentiation | 5 | 1.37 | -1.05 | 1.2 | -1.12 | 1.36 | 2.17 | 1.48 |
| peripheral nervous system neuron development | 5 | 1.37 | -1.05 | 1.2 | -1.12 | 1.36 | 2.17 | 1.48 |
| m7G(5')pppN diphosphatase activity | 2 | -1.05 | 1.25 | 1.14 | 1.39 | -1.04 | 1.03 | 1.06 |
| inositol oxygenase activity | 1 | 3.92 | -4.04 | -3.69 | -3.19 | -2.14 | 5.76 | -2.59 |
| NADH dehydrogenase (quinone) activity | 30 | 1.12 | 1.03 | 1.27 | 1.07 | 1.1 | 1.73 | -1.3 |
| rhodopsin kinase activity | 1 | 2.38 | 1.14 | 1.51 | 1.03 | 1.36 | 1.67 | 1.26 |
| sphingomyelin phosphodiesterase D activity | 1 | -1.04 | 1.38 | 1.42 | 1.29 | 1.65 | 1.05 | 1.22 |
| sugar-phosphatase activity | 3 | 1.95 | -1.01 | 1.52 | -1.38 | -1.21 | 1.92 | 1.24 |
| sugar-terminal-phosphatase activity | 1 | 1.02 | 1.09 | 1.02 | 1.0 | -1.15 | -1.1 | -1.14 |
| tau-protein kinase activity | 1 | 1.73 | 1.2 | 1.66 | 1.31 | 1.35 | 4.88 | 1.77 |
| testosterone 17-beta-dehydrogenase (NAD+) activity | 1 | 1.03 | -1.04 | 1.2 | -1.01 | 1.0 | 2.11 | -1.07 |
| trimethyllysine dioxygenase activity | 1 | 1.3 | -1.08 | -1.07 | -1.13 | -1.19 | 1.96 | -1.06 |
| transferase activity, transferring alkylthio groups | 1 | 1.08 | 1.05 | 1.26 | 1.07 | 1.31 | 1.33 | 1.1 |
| glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity | 2 | 1.46 | 1.48 | 1.74 | 1.25 | 2.09 | 1.65 | 1.44 |
| N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity | 2 | 1.46 | 1.48 | 1.74 | 1.25 | 2.09 | 1.65 | 1.44 |
| coenzyme-B sulfoethylthiotransferase activity | 1 | 1.08 | 1.05 | 1.26 | 1.07 | 1.31 | 1.33 | 1.1 |
| glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity | 3 | 2.47 | -1.53 | -1.66 | -1.5 | -1.37 | 1.3 | 1.19 |
| GDP-L-fucose synthase activity | 1 | 1.27 | 1.21 | 1.12 | 1.11 | 1.24 | -1.28 | 1.03 |
| chondroitin sulfate proteoglycan biosynthetic process | 7 | 1.31 | 1.13 | 1.16 | 1.13 | 1.24 | 1.3 | 1.08 |
| chondroitin sulfate proteoglycan metabolic process | 7 | 1.31 | 1.13 | 1.16 | 1.13 | 1.24 | 1.3 | 1.08 |
| nucleic acid transport | 47 | -1.07 | 1.16 | 1.25 | 1.13 | 1.27 | 1.15 | 1.02 |
| RNA transport | 47 | -1.07 | 1.16 | 1.25 | 1.13 | 1.27 | 1.15 | 1.02 |
| flavin adenine dinucleotide binding | 60 | 1.01 | 1.02 | -1.23 | 1.0 | 1.01 | -1.05 | 1.0 |
| NADP binding | 12 | 1.43 | 1.07 | -1.48 | -1.04 | -1.1 | 1.24 | 1.21 |
| coenzyme binding | 127 | -1.03 | 1.05 | -1.16 | 1.03 | 1.04 | -1.04 | -1.01 |
| oxidoreductase activity, acting on NADH or NADPH, oxygen as acceptor | 1 | 2.71 | -1.01 | 1.01 | -1.06 | -1.03 | 2.12 | 1.84 |
| epithelial cell proliferation | 11 | 1.14 | 1.03 | 1.13 | -1.0 | 1.25 | 1.04 | 1.44 |
| regulation of epithelial cell proliferation | 2 | 1.23 | 1.09 | 1.59 | 1.34 | 1.35 | -1.03 | -1.14 |
| negative regulation of epithelial cell proliferation | 1 | 1.14 | 1.03 | 1.49 | 1.35 | 1.3 | -1.2 | -1.41 |
| regulation of mRNA processing | 63 | 1.2 | 1.17 | 1.28 | 1.16 | 1.23 | 1.4 | 1.09 |
| positive regulation of mRNA processing | 4 | 1.18 | 1.11 | 1.21 | 1.11 | 1.04 | 1.21 | 1.05 |
| negative regulation of mRNA processing | 4 | 1.46 | 1.2 | 1.32 | 1.17 | 1.27 | 1.62 | 1.09 |
| regulation of defense response to virus | 2 | 1.7 | 1.18 | 1.38 | 1.15 | 1.21 | 1.1 | 1.08 |
| regulation of protein secretion | 7 | -1.08 | 1.14 | 1.13 | 1.04 | -1.11 | -1.14 | 1.11 |
| negative regulation of protein secretion | 2 | -1.13 | 1.08 | -1.08 | -1.02 | -1.24 | -1.25 | -1.14 |
| positive regulation of protein secretion | 4 | 1.08 | 1.1 | 1.36 | 1.08 | 1.05 | 1.04 | 1.2 |
| regulation of peptidyl-tyrosine phosphorylation | 3 | 1.04 | 1.16 | 1.54 | 1.06 | 1.26 | 1.21 | 1.03 |
| negative regulation of peptidyl-tyrosine phosphorylation | 2 | -1.04 | 1.11 | 1.79 | -1.0 | 1.29 | 1.24 | 1.01 |
| low-density lipoprotein particle receptor binding | 1 | 3.66 | -1.09 | 1.54 | -1.1 | 1.65 | 2.15 | 1.33 |
| regulation of phagocytosis | 1 | 2.18 | -2.11 | 2.19 | 1.49 | 1.48 | 1.37 | 1.38 |
| regulation of neurogenesis | 95 | 1.06 | 1.12 | 1.34 | 1.07 | 1.33 | 1.21 | 1.16 |
| negative regulation of neurogenesis | 22 | -1.0 | 1.18 | 1.36 | 1.09 | 1.44 | 1.27 | 1.22 |
| positive regulation of neurogenesis | 5 | 1.42 | 1.13 | 1.48 | 1.05 | 1.3 | 1.35 | 1.31 |
| regulation of axonogenesis | 27 | -1.08 | 1.16 | 1.5 | 1.05 | 1.39 | 1.19 | 1.15 |
| negative regulation of axonogenesis | 5 | -1.24 | 1.29 | 1.61 | 1.16 | 1.56 | -1.15 | 1.13 |
| regulation of dendrite development | 22 | 1.22 | 1.13 | 1.27 | 1.1 | 1.38 | 1.38 | 1.24 |
| negative regulation of dendrite morphogenesis | 5 | 1.32 | 1.12 | 1.2 | -1.01 | 1.18 | 1.31 | 1.53 |
| positive regulation of dendrite morphogenesis | 1 | 1.25 | 1.26 | 1.64 | 1.09 | 1.9 | 1.54 | 1.83 |
| regulation of immune response | 48 | 1.38 | -1.08 | 1.02 | -1.04 | 1.04 | 1.2 | 1.15 |
| negative regulation of immune response | 7 | 1.96 | -1.13 | 1.13 | -1.03 | 1.24 | 1.15 | 1.14 |
| positive regulation of immune response | 6 | 1.81 | 1.02 | 1.25 | 1.0 | 1.15 | 1.79 | 1.27 |
| regulation of biological process | 2435 | 1.15 | 1.06 | 1.19 | 1.05 | 1.18 | 1.18 | 1.12 |
| regulation of catalytic activity | 148 | 1.21 | 1.11 | 1.27 | 1.09 | 1.25 | 1.25 | 1.16 |
| regulation of viral reproduction | 4 | 1.27 | 1.23 | 1.52 | 1.17 | 1.6 | 1.47 | 1.34 |
| regulation of developmental process | 392 | 1.17 | 1.08 | 1.27 | 1.05 | 1.26 | 1.24 | 1.19 |
| regulation of cellular process | 2258 | 1.15 | 1.06 | 1.19 | 1.05 | 1.19 | 1.18 | 1.13 |
| regulation of behavior | 32 | -1.08 | 1.03 | 1.08 | -1.14 | 1.06 | 1.06 | 1.14 |
| thymidylate synthase (FAD) activity | 1 | 1.78 | 1.14 | -1.44 | 1.08 | 1.0 | 3.89 | 1.83 |
| circadian sleep/wake cycle, sleep | 1 | -1.57 | 1.18 | 1.13 | 1.01 | 1.08 | -1.1 | 1.28 |
| regulation of synapse structure and activity | 62 | 1.04 | 1.11 | 1.34 | 1.06 | 1.23 | 1.18 | 1.19 |
| regulation of synaptic transmission | 36 | 1.13 | 1.11 | 1.3 | 1.02 | 1.17 | 1.29 | 1.22 |
| negative regulation of synaptic transmission | 2 | -1.06 | 1.07 | 1.19 | 1.01 | 1.07 | 1.19 | 1.07 |
| positive regulation of synaptic transmission | 2 | -1.81 | 1.29 | 2.08 | -1.04 | 1.39 | 1.52 | 1.29 |
| regulation of synapse organization | 47 | -1.01 | 1.12 | 1.34 | 1.09 | 1.27 | 1.13 | 1.21 |
| synapse organization | 72 | 1.22 | 1.16 | 1.24 | 1.06 | 1.23 | 1.32 | 1.22 |
| coagulation | 5 | 1.95 | -1.51 | -1.67 | -1.6 | -1.41 | 1.79 | -1.02 |
| regulation of body fluid levels | 10 | 1.04 | -1.31 | -1.24 | -1.25 | -1.09 | -1.06 | -1.17 |
| multicellular organismal movement | 2 | -1.89 | -1.52 | -1.42 | -1.43 | -1.62 | -1.61 | 2.67 |
| musculoskeletal movement | 2 | -1.89 | -1.52 | -1.42 | -1.43 | -1.62 | -1.61 | 2.67 |
| cognition | 104 | 1.16 | 1.04 | 1.18 | -1.01 | 1.23 | 1.38 | 1.22 |
| response to stimulus | 1939 | 1.15 | 1.01 | 1.09 | -1.01 | 1.12 | 1.16 | 1.12 |
| cobalt ion binding | 1 | 2.85 | -1.01 | 1.16 | 1.03 | -1.08 | 1.74 | -1.1 |
| neuromuscular process | 14 | 1.16 | 1.16 | 1.49 | 1.02 | 1.5 | 1.39 | 1.36 |
| detection of stimulus involved in sensory perception | 56 | 1.14 | -1.01 | 1.02 | 1.02 | 1.07 | 1.15 | 1.02 |
| detection of chemical stimulus involved in sensory perception | 31 | 1.02 | -1.01 | -1.04 | -1.0 | 1.05 | 1.09 | -1.03 |
| detection of light stimulus involved in visual perception | 19 | 1.33 | 1.01 | 1.14 | 1.06 | 1.12 | 1.18 | 1.06 |
| detection of chemical stimulus involved in sensory perception of smell | 15 | -1.07 | -1.01 | -1.04 | 1.01 | -1.07 | 1.07 | -1.07 |
| sensory perception of bitter taste | 2 | -1.31 | -1.2 | -1.22 | -1.24 | 3.17 | 1.22 | -1.17 |
| sensory perception of salty taste | 2 | 1.15 | 1.12 | 1.11 | 1.03 | -1.03 | 1.1 | -1.05 |
| sensory perception of sweet taste | 10 | 1.42 | 1.09 | 1.21 | 1.04 | 1.18 | 1.17 | 1.2 |
| positive chemotaxis | 5 | 1.16 | -1.25 | 1.29 | -1.0 | 1.37 | 2.36 | -1.31 |
| negative chemotaxis | 4 | -1.03 | -1.01 | 1.19 | 1.16 | 1.89 | -1.4 | -1.09 |
| regulation of chemotaxis | 5 | 1.06 | 1.07 | 1.36 | 1.03 | 1.13 | 1.06 | 1.02 |
| negative regulation of chemotaxis | 1 | 1.49 | 1.07 | 1.15 | 1.04 | -1.03 | 1.53 | -1.09 |
| pigment cell differentiation | 1 | 1.43 | 1.13 | 3.64 | 1.22 | 1.75 | 1.43 | 1.9 |
| sensory perception of temperature stimulus | 2 | 1.0 | 1.06 | -1.05 | 1.0 | -1.03 | -1.07 | -1.17 |
| sensory perception of light stimulus | 41 | 1.14 | -1.01 | 1.16 | 1.08 | 1.09 | 1.11 | 1.02 |
| thermoception | 2 | 1.0 | 1.06 | -1.05 | 1.0 | -1.03 | -1.07 | -1.17 |
| magnetoreception | 1 | 1.37 | -1.11 | -1.04 | 1.15 | 1.03 | 2.71 | 1.13 |
| detection of temperature stimulus involved in thermoception | 2 | 1.0 | 1.06 | -1.05 | 1.0 | -1.03 | -1.07 | -1.17 |
| detection of temperature stimulus involved in sensory perception | 2 | 1.0 | 1.06 | -1.05 | 1.0 | -1.03 | -1.07 | -1.17 |
| detection of light stimulus involved in sensory perception | 20 | 1.33 | 1.01 | 1.13 | 1.06 | 1.11 | 1.23 | 1.07 |
| detection of mechanical stimulus involved in sensory perception | 3 | 1.34 | -1.14 | 1.07 | -1.03 | 1.11 | 1.5 | 1.37 |
| detection of light stimulus involved in magnetoreception | 1 | 1.37 | -1.11 | -1.04 | 1.15 | 1.03 | 2.71 | 1.13 |
| detection of mechanical stimulus | 3 | 1.34 | -1.14 | 1.07 | -1.03 | 1.11 | 1.5 | 1.37 |
| regulation of lipid catabolic process | 1 | -3.9 | 1.39 | 1.21 | 1.36 | 1.18 | -2.17 | -1.43 |
| negative regulation of lipid catabolic process | 1 | -3.9 | 1.39 | 1.21 | 1.36 | 1.18 | -2.17 | -1.43 |
| regulation of nitric-oxide synthase activity | 1 | 1.08 | 1.16 | -1.5 | 1.55 | 1.01 | -2.04 | -1.2 |
| spindle midzone | 11 | -1.06 | 1.06 | 1.11 | 1.22 | 1.35 | -1.12 | -1.06 |
| negative regulation of nitric-oxide synthase activity | 1 | 1.08 | 1.16 | -1.5 | 1.55 | 1.01 | -2.04 | -1.2 |
| microtubule plus-end binding | 1 | 1.58 | -1.1 | -1.13 | -1.09 | -1.05 | -1.25 | 1.03 |
| microtubule minus-end binding | 3 | 2.17 | 1.08 | 1.62 | 1.15 | 1.7 | 2.3 | 1.31 |
| microtubule sliding | 2 | -1.0 | 1.03 | 1.08 | 1.16 | 1.45 | -1.14 | -1.41 |
| GTPase binding | 33 | 1.09 | 1.2 | 1.54 | 1.1 | 1.4 | 1.28 | 1.4 |
| mRNA transport | 21 | 1.04 | 1.14 | 1.2 | 1.17 | 1.19 | 1.11 | -1.01 |
| snRNA transport | 2 | 1.9 | 1.18 | 1.33 | 1.29 | 1.62 | 1.23 | 1.13 |
| tRNA transport | 1 | 1.48 | 1.1 | 1.33 | 1.32 | 1.54 | -1.18 | -1.04 |
| regulation of endosome size | 4 | 1.23 | 1.04 | 1.25 | 1.26 | 1.24 | -1.0 | -1.18 |
| regulation of transcription during meiosis | 3 | 1.04 | 1.04 | 1.22 | 1.08 | 1.06 | -1.01 | -1.03 |
| positive regulation of transcription during meiosis | 3 | 1.04 | 1.04 | 1.22 | 1.08 | 1.06 | -1.01 | -1.03 |
| regulation of secretion | 26 | 1.02 | 1.09 | 1.15 | 1.04 | 1.03 | 1.23 | 1.1 |
| positive regulation of secretion | 9 | -1.15 | 1.19 | 1.43 | 1.11 | 1.18 | 1.14 | 1.14 |
| negative regulation of secretion | 3 | -1.01 | 1.14 | -1.02 | 1.03 | -1.12 | -1.05 | -1.01 |
| regulation of transport | 97 | 1.11 | 1.07 | 1.23 | 1.06 | 1.15 | 1.32 | 1.13 |
| positive regulation of transport | 43 | 1.03 | 1.09 | 1.27 | 1.08 | 1.22 | 1.17 | 1.12 |
| negative regulation of transport | 13 | 1.41 | 1.11 | 1.22 | 1.06 | 1.16 | 1.64 | 1.33 |
| regulation of DNA metabolic process | 23 | -1.05 | 1.12 | 1.21 | 1.14 | 1.34 | -1.12 | 1.07 |
| negative regulation of DNA metabolic process | 12 | -1.26 | 1.11 | 1.2 | 1.14 | 1.37 | -1.28 | 1.13 |
| positive regulation of DNA metabolic process | 4 | 1.13 | 1.17 | 1.08 | 1.14 | 1.28 | -1.01 | 1.07 |
| negative regulation of lipid biosynthetic process | 1 | -84.21 | -3.59 | 1.42 | 2.07 | 1.14 | -58.36 | -15.07 |
| regulation of small GTPase mediated signal transduction | 88 | 1.04 | 1.15 | 1.37 | 1.03 | 1.3 | 1.29 | 1.25 |
| positive regulation of small GTPase mediated signal transduction | 2 | 1.82 | 1.14 | 1.61 | 1.13 | 1.83 | 2.54 | 1.37 |
| negative regulation of small GTPase mediated signal transduction | 7 | -1.35 | 1.4 | 1.44 | 1.09 | 1.53 | 1.07 | 1.44 |
| protein tetramerization | 3 | 1.61 | 1.24 | 1.33 | 1.08 | 1.47 | 1.59 | 1.11 |
| meiotic nuclear envelope disassembly | 2 | 1.15 | 1.34 | 1.04 | 1.15 | 1.01 | -1.24 | 1.13 |
| nuclear envelope disassembly | 2 | 1.15 | 1.34 | 1.04 | 1.15 | 1.01 | -1.24 | 1.13 |
| 'de novo' posttranslational protein folding | 2 | 1.05 | 1.15 | 1.39 | 1.19 | 1.32 | -1.27 | 1.18 |
| chaperone mediated protein folding requiring cofactor | 2 | 1.05 | 1.15 | 1.39 | 1.19 | 1.32 | -1.27 | 1.18 |
| regulation of sequence-specific DNA binding transcription factor activity | 13 | 1.34 | 1.18 | 1.37 | 1.07 | 1.27 | 1.33 | 1.33 |
| positive regulation of sequence-specific DNA binding transcription factor activity | 8 | 1.24 | 1.17 | 1.52 | 1.19 | 1.56 | 1.5 | 1.11 |
| positive regulation of NF-kappaB transcription factor activity | 2 | 1.33 | 1.09 | 1.29 | 1.43 | 1.67 | 1.2 | -1.13 |
| negative regulation of developmental process | 122 | 1.1 | 1.09 | 1.28 | 1.07 | 1.29 | 1.11 | 1.17 |
| positive regulation of developmental process | 44 | 1.22 | 1.09 | 1.22 | 1.05 | 1.23 | 1.18 | 1.21 |
| regulation of binding | 11 | 1.19 | 1.27 | 1.43 | 1.19 | 1.41 | 1.05 | 1.28 |
| positive regulation of binding | 1 | 2.81 | 1.45 | 1.89 | 1.09 | 1.48 | 2.58 | 1.71 |
| negative regulation of binding | 3 | 1.1 | 1.28 | 1.83 | 1.03 | 1.28 | 1.14 | 1.67 |
| regulation of DNA binding | 7 | 1.27 | 1.36 | 1.52 | 1.13 | 1.3 | 1.15 | 1.52 |
| DNA ligation involved in DNA repair | 2 | 1.6 | 1.14 | 1.36 | 1.04 | 1.35 | 1.44 | 1.13 |
| synaptic growth at neuromuscular junction | 18 | 1.09 | 1.23 | 1.45 | 1.08 | 1.28 | 1.56 | 1.28 |
| regulation of cellular component organization | 367 | 1.14 | 1.11 | 1.34 | 1.08 | 1.34 | 1.31 | 1.22 |
| negative regulation of cellular component organization | 63 | 1.04 | 1.14 | 1.37 | 1.11 | 1.35 | 1.19 | 1.15 |
| positive regulation of cellular component organization | 47 | 1.11 | 1.11 | 1.32 | 1.07 | 1.32 | 1.16 | 1.27 |
| striated muscle cell differentiation | 106 | 1.33 | 1.05 | 1.26 | -1.02 | 1.19 | 1.4 | 1.16 |
| regulation of muscle cell differentiation | 40 | 1.03 | 1.1 | 1.31 | 1.08 | 1.23 | 1.12 | 1.2 |
| regulation of striated muscle cell differentiation | 40 | 1.03 | 1.1 | 1.31 | 1.08 | 1.23 | 1.12 | 1.2 |
| nuclear export | 29 | 1.21 | 1.09 | 1.3 | 1.16 | 1.26 | 1.19 | 1.04 |
| nuclear transport | 70 | 1.17 | 1.16 | 1.34 | 1.19 | 1.33 | 1.24 | 1.11 |
| nuclear import | 37 | 1.11 | 1.21 | 1.35 | 1.2 | 1.36 | 1.25 | 1.16 |
| regulation of nitrogen compound metabolic process | 800 | 1.13 | 1.09 | 1.18 | 1.07 | 1.17 | 1.17 | 1.12 |
| negative regulation of nitrogen compound metabolic process | 199 | 1.16 | 1.09 | 1.16 | 1.05 | 1.22 | 1.21 | 1.14 |
| positive regulation of nitrogen compound metabolic process | 222 | 1.11 | 1.05 | 1.13 | 1.03 | 1.13 | 1.18 | 1.15 |
| regulation of phosphorus metabolic process | 80 | 1.29 | 1.13 | 1.33 | 1.09 | 1.29 | 1.35 | 1.2 |
| meiotic sister chromatid cohesion | 4 | -1.07 | 1.07 | 1.03 | 1.03 | 1.12 | -1.1 | -1.1 |
| coenzyme transporter activity | 1 | -1.28 | 1.24 | -1.24 | 1.18 | -1.13 | -1.69 | 1.59 |
| cofactor metabolic process | 102 | 1.31 | 1.1 | 1.11 | 1.03 | 1.09 | 1.32 | 1.04 |
| cofactor catabolic process | 38 | 1.01 | 1.1 | 1.28 | -1.04 | 1.08 | 1.52 | -1.03 |
| cofactor biosynthetic process | 53 | 1.51 | 1.12 | 1.11 | 1.08 | 1.1 | 1.22 | 1.09 |
| protein insertion into membrane | 2 | -1.2 | 1.31 | 1.29 | 1.27 | 1.21 | 1.76 | -1.19 |
| release of sequestered calcium ion into cytosol | 1 | 2.52 | 1.29 | 3.07 | -1.16 | 1.37 | 5.13 | 2.97 |
| dioxygenase activity | 52 | 1.01 | 1.03 | -1.04 | 1.03 | 1.01 | 1.06 | -1.01 |
| phosphoprotein binding | 3 | 1.02 | 1.34 | 1.39 | 1.03 | 1.34 | 1.5 | 1.35 |
| cytoplasmic sequestering of protein | 4 | 2.74 | 1.08 | 1.33 | 1.02 | 1.2 | 2.19 | 1.75 |
| positive regulation of protein transport | 25 | 1.11 | 1.07 | 1.24 | 1.09 | 1.26 | 1.13 | 1.14 |
| regulation of protein transport | 40 | 1.14 | 1.11 | 1.23 | 1.09 | 1.2 | 1.19 | 1.2 |
| negative regulation of protein transport | 8 | 1.65 | 1.12 | 1.19 | 1.05 | 1.14 | 1.51 | 1.37 |
| spindle assembly | 37 | 1.19 | 1.02 | 1.18 | -1.02 | 1.27 | 1.27 | 1.15 |
| spindle elongation | 74 | 1.39 | 1.08 | 1.2 | 1.05 | 1.24 | 1.22 | 1.15 |
| establishment of localization | 1375 | -1.06 | 1.01 | 1.01 | -1.0 | 1.07 | 1.08 | 1.01 |
| maintenance of location | 51 | 1.21 | 1.11 | 1.23 | 1.06 | 1.24 | 1.26 | 1.22 |
| establishment of RNA localization | 48 | -1.05 | 1.16 | 1.27 | 1.13 | 1.3 | 1.18 | 1.03 |
| maintenance of RNA location | 3 | 1.53 | 1.04 | 1.01 | 1.11 | 1.01 | -1.06 | -1.06 |
| sequestering of metal ion | 1 | 1.04 | 1.04 | -1.04 | -1.09 | -1.16 | -1.14 | -1.1 |
| regulation of multicellular organismal process | 386 | 1.12 | 1.08 | 1.22 | 1.02 | 1.2 | 1.23 | 1.19 |
| positive regulation of multicellular organismal process | 31 | 1.01 | 1.17 | 1.34 | 1.03 | 1.25 | 1.29 | 1.33 |
| negative regulation of multicellular organismal process | 14 | 1.3 | 1.06 | 1.12 | -1.04 | 1.17 | 1.15 | 1.14 |
| regulation of protein metabolic process | 182 | 1.3 | 1.11 | 1.31 | 1.09 | 1.29 | 1.35 | 1.22 |
| positive regulation of protein metabolic process | 25 | 1.12 | 1.15 | 1.46 | 1.08 | 1.34 | 1.24 | 1.28 |
| negative regulation of protein metabolic process | 47 | 1.33 | 1.04 | 1.25 | 1.02 | 1.25 | 1.33 | 1.24 |
| regulation of RNA metabolic process | 733 | 1.14 | 1.09 | 1.18 | 1.07 | 1.17 | 1.17 | 1.12 |
| negative regulation of RNA metabolic process | 190 | 1.17 | 1.09 | 1.17 | 1.04 | 1.21 | 1.23 | 1.16 |
| positive regulation of RNA metabolic process | 213 | 1.11 | 1.05 | 1.13 | 1.03 | 1.13 | 1.19 | 1.15 |
| spindle midzone assembly | 2 | -1.32 | -1.05 | -1.11 | 1.02 | 1.0 | -1.24 | -1.14 |
| spindle midzone assembly involved in meiosis | 2 | -1.32 | -1.05 | -1.11 | 1.02 | 1.0 | -1.24 | -1.14 |
| protein polymerization | 21 | -1.19 | 1.04 | 1.18 | 1.11 | 1.2 | -1.18 | -1.09 |
| protein oligomerization | 7 | 1.05 | 1.23 | 1.4 | 1.02 | 1.36 | 1.37 | 1.51 |
| protein depolymerization | 9 | -1.16 | 1.13 | 1.31 | 1.1 | 1.39 | -1.02 | -1.01 |
| regulation of cellular component movement | 12 | 1.86 | -1.05 | 1.08 | -1.07 | 1.19 | 1.24 | 1.56 |
| negative regulation of cellular component movement | 2 | 2.1 | 1.1 | 1.42 | -1.08 | 1.39 | 1.72 | 1.26 |
| regulation of sequestering of calcium ion | 2 | 1.38 | 1.1 | 1.6 | 1.13 | 1.03 | 2.02 | 1.52 |
| negative regulation of sequestering of calcium ion | 2 | 1.38 | 1.1 | 1.6 | 1.13 | 1.03 | 2.02 | 1.52 |
| cell tip | 1 | 3.87 | -1.1 | 1.38 | 1.1 | 2.2 | 1.95 | 1.38 |
| NAD binding | 36 | 1.03 | 1.15 | -1.0 | -1.0 | -1.02 | 1.24 | 1.11 |
| protein homotetramerization | 2 | 1.92 | 1.2 | 1.42 | 1.06 | 1.79 | 1.71 | 1.14 |
| protein heterotetramerization | 1 | 1.14 | 1.34 | 1.17 | 1.13 | 1.0 | 1.38 | 1.04 |
| protein heterooligomerization | 1 | 1.14 | 1.34 | 1.17 | 1.13 | 1.0 | 1.38 | 1.04 |
| establishment of spindle localization | 15 | -1.09 | 1.17 | 1.26 | 1.1 | 1.29 | 1.1 | 1.08 |
| establishment of spindle orientation | 7 | 1.09 | 1.1 | 1.4 | 1.07 | 1.32 | 1.26 | 1.14 |
| establishment of meiotic spindle localization | 2 | 1.18 | 1.0 | 1.04 | 1.03 | 1.06 | -1.17 | 1.01 |
| establishment of meiotic spindle orientation | 1 | 1.58 | -1.1 | -1.13 | -1.09 | -1.05 | -1.25 | 1.03 |
| centrosome organization | 107 | 1.17 | 1.09 | 1.19 | 1.09 | 1.25 | 1.12 | 1.07 |
| centrosome duplication | 77 | 1.18 | 1.09 | 1.19 | 1.07 | 1.26 | 1.14 | 1.09 |
| centrosome separation | 12 | 1.08 | 1.11 | 1.11 | 1.12 | 1.23 | 1.13 | 1.05 |
| cell division | 175 | 1.03 | 1.09 | 1.25 | 1.09 | 1.33 | 1.06 | 1.09 |
| regulation of cell division | 6 | -1.42 | 1.36 | 1.46 | 1.12 | 1.27 | -1.11 | 1.24 |
| establishment of chromosome localization | 18 | 1.21 | 1.05 | 1.2 | 1.17 | 1.42 | 1.12 | -1.0 |
| chromosome separation | 9 | 1.18 | 1.08 | 1.24 | 1.13 | 1.49 | 1.15 | 1.05 |
| chromosome movement towards spindle pole | 2 | 1.27 | 1.03 | -1.05 | 1.3 | 1.32 | -1.33 | -1.18 |
| mitotic sister chromatid separation | 5 | 1.35 | 1.08 | 1.24 | 1.1 | 1.53 | 1.17 | 1.04 |
| meiotic chromosome separation | 4 | 1.0 | 1.09 | 1.25 | 1.17 | 1.45 | 1.14 | 1.06 |
| metaphase plate congression | 17 | 1.19 | 1.05 | 1.22 | 1.17 | 1.43 | 1.15 | 1.01 |
| meiotic metaphase plate congression | 1 | -1.17 | 1.02 | -1.13 | 1.11 | -1.02 | -1.2 | -1.13 |
| attachment of spindle microtubules to chromosome | 5 | -1.0 | 1.08 | 1.05 | 1.15 | 1.21 | 1.0 | 1.02 |
| attachment of spindle microtubules to kinetochore involved in mitotic sister chromatid segregation | 1 | -1.05 | 1.32 | 1.38 | -1.11 | 1.3 | 2.36 | 1.79 |
| attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation | 1 | -1.24 | -1.06 | -1.4 | -1.21 | -1.33 | -1.23 | -1.12 |
| G1 phase | 2 | 1.34 | 1.36 | 1.26 | 1.22 | 1.44 | 1.51 | 1.45 |
| G2 phase | 1 | 1.67 | 1.26 | 1.32 | 1.4 | 1.23 | 1.13 | 1.21 |
| S phase | 4 | 1.02 | 1.08 | 1.18 | 1.25 | 1.3 | 1.01 | -1.14 |
| meiotic cell cycle | 132 | 1.13 | 1.09 | 1.16 | 1.08 | 1.21 | 1.1 | 1.09 |
| anaphase | 25 | 1.25 | 1.11 | 1.26 | 1.14 | 1.31 | 1.3 | 1.1 |
| metaphase | 2 | 1.14 | 1.11 | 1.14 | 1.15 | 1.24 | 1.02 | -1.01 |
| prophase | 3 | 1.29 | 1.04 | 1.3 | -1.01 | 1.37 | 1.56 | 1.26 |
| interphase | 85 | 1.24 | 1.15 | 1.34 | 1.2 | 1.41 | 1.23 | 1.13 |
| telophase | 1 | 1.4 | 1.02 | 1.15 | 1.14 | 1.72 | -1.18 | -1.16 |
| M phase of meiotic cell cycle | 124 | 1.1 | 1.11 | 1.17 | 1.11 | 1.22 | 1.09 | 1.08 |
| interphase of meiotic cell cycle | 1 | 1.43 | -1.05 | 1.72 | 1.27 | 1.65 | 1.43 | -1.28 |
| interphase of mitotic cell cycle | 84 | 1.23 | 1.15 | 1.33 | 1.2 | 1.41 | 1.23 | 1.14 |
| S phase of meiotic cell cycle | 1 | 1.43 | -1.05 | 1.72 | 1.27 | 1.65 | 1.43 | -1.28 |
| regulation of hydrolase activity | 77 | 1.13 | 1.1 | 1.24 | 1.07 | 1.24 | 1.19 | 1.15 |
| regulation of transferase activity | 51 | 1.28 | 1.15 | 1.34 | 1.13 | 1.31 | 1.38 | 1.19 |
| regulation of lyase activity | 8 | 1.21 | 1.07 | 1.14 | -1.02 | 1.05 | 1.21 | -1.04 |
| regulation of ligase activity | 5 | 1.5 | 1.1 | 1.38 | 1.2 | 1.15 | -1.03 | 1.17 |
| regulation of oxidoreductase activity | 2 | 1.57 | 1.18 | -1.18 | 1.24 | 1.17 | 1.04 | 1.44 |
| positive regulation of hydrolase activity | 26 | 1.05 | 1.14 | 1.29 | 1.16 | 1.4 | 1.23 | 1.08 |
| negative regulation of hydrolase activity | 6 | -1.1 | 1.0 | 1.21 | 1.14 | 1.15 | -1.28 | -1.18 |
| positive regulation of transferase activity | 25 | 1.18 | 1.14 | 1.29 | 1.1 | 1.23 | 1.49 | 1.19 |
| negative regulation of transferase activity | 12 | 1.53 | 1.14 | 1.46 | 1.09 | 1.41 | 1.69 | 1.32 |
| positive regulation of lyase activity | 7 | 1.18 | 1.07 | 1.16 | -1.04 | 1.05 | 1.21 | -1.04 |
| negative regulation of lyase activity | 1 | 1.44 | 1.03 | -1.0 | 1.12 | 1.06 | 1.16 | -1.02 |
| positive regulation of ligase activity | 2 | -1.3 | 1.19 | -1.18 | 1.16 | -1.1 | -2.07 | -1.12 |
| negative regulation of ligase activity | 1 | 1.67 | 1.26 | 1.32 | 1.4 | 1.23 | 1.13 | 1.21 |
| positive regulation of oxidoreductase activity | 1 | 2.28 | 1.19 | 1.07 | -1.0 | 1.37 | 2.2 | 2.5 |
| negative regulation of oxidoreductase activity | 1 | 1.08 | 1.16 | -1.5 | 1.55 | 1.01 | -2.04 | -1.2 |
| kinetochore assembly | 3 | 1.0 | 1.03 | -1.07 | 1.16 | 1.05 | -1.12 | -1.1 |
| kinetochore organization | 7 | -1.05 | 1.08 | 1.04 | 1.13 | 1.18 | 1.01 | 1.0 |
| alpha-actinin binding | 1 | 1.14 | -1.01 | 1.12 | 1.41 | 1.03 | -1.2 | -1.43 |
| neuron apoptotic process | 2 | -1.07 | 1.32 | 1.73 | -1.0 | -1.14 | 1.12 | 1.39 |
| stress-activated MAPK cascade | 26 | -1.01 | 1.14 | 1.45 | 1.09 | 1.5 | 1.22 | 1.4 |
| hormone receptor binding | 5 | 1.32 | -1.42 | -1.36 | -1.38 | -1.32 | -1.27 | 1.43 |
| peptide hormone receptor binding | 1 | 2.14 | -13.11 | -13.63 | -12.76 | -16.87 | -14.38 | 5.61 |
| negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 1 | 1.67 | 1.26 | 1.32 | 1.4 | 1.23 | 1.13 | 1.21 |
| regulation of ubiquitin-protein ligase activity | 4 | 1.88 | 1.03 | 1.57 | 1.14 | 1.19 | 1.34 | 1.27 |
| regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 1 | 1.67 | 1.26 | 1.32 | 1.4 | 1.23 | 1.13 | 1.21 |
| regulation of ubiquitin-protein ligase activity involved in meiotic cell cycle | 1 | 1.0 | -1.01 | -1.14 | -1.09 | -1.19 | -1.12 | -1.06 |
| positive regulation of ubiquitin-protein ligase activity involved in meiotic cell cycle | 1 | 1.0 | -1.01 | -1.14 | -1.09 | -1.19 | -1.12 | -1.06 |
| positive regulation of ubiquitin-protein ligase activity | 1 | 1.0 | -1.01 | -1.14 | -1.09 | -1.19 | -1.12 | -1.06 |
| negative regulation of ubiquitin-protein ligase activity | 1 | 1.67 | 1.26 | 1.32 | 1.4 | 1.23 | 1.13 | 1.21 |
| regulation of meiotic cell cycle | 11 | 1.37 | 1.1 | 1.22 | 1.11 | 1.24 | 1.39 | 1.14 |
| myoblast proliferation | 3 | -1.05 | 1.1 | 1.17 | 1.05 | 1.18 | 1.15 | 1.02 |
| intracellular pH reduction | 6 | -1.56 | 1.28 | 1.81 | 1.0 | 1.29 | 1.81 | 1.32 |
| regulation of intracellular pH | 6 | -1.56 | 1.28 | 1.81 | 1.0 | 1.29 | 1.81 | 1.32 |
| maintenance of protein location in nucleus | 1 | 1.16 | 1.03 | 1.19 | 1.34 | 1.67 | -1.1 | 1.01 |
| cytosolic calcium ion homeostasis | 4 | 1.44 | 1.13 | 1.5 | -1.03 | 1.13 | 1.91 | 1.33 |
| elevation of cytosolic calcium ion concentration involved in G-protein signaling coupled to IP3 second messenger | 2 | 1.11 | 1.1 | 1.07 | 1.07 | 1.09 | 1.04 | 1.11 |
| activation of anaphase-promoting complex activity involved in meiotic cell cycle | 1 | 1.0 | -1.01 | -1.14 | -1.09 | -1.19 | -1.12 | -1.06 |
| activation of anaphase-promoting complex activity | 1 | 1.0 | -1.01 | -1.14 | -1.09 | -1.19 | -1.12 | -1.06 |
| regulation of filopodium assembly | 9 | 1.19 | 1.09 | 1.6 | 1.1 | 1.48 | -1.13 | 1.2 |
| negative regulation of filopodium assembly | 2 | -1.38 | -1.03 | 2.18 | 1.01 | 1.59 | -1.39 | -1.04 |
| positive regulation of filopodium assembly | 5 | 1.14 | 1.08 | 1.54 | 1.11 | 1.41 | -1.3 | 1.15 |
| regulation of cytoskeleton organization | 60 | -1.04 | 1.1 | 1.42 | 1.04 | 1.42 | 1.16 | 1.22 |
| negative regulation of cytoskeleton organization | 17 | 1.02 | 1.13 | 1.55 | 1.04 | 1.43 | 1.45 | 1.22 |
| positive regulation of cytoskeleton organization | 10 | 1.03 | 1.13 | 1.32 | -1.01 | 1.32 | 1.08 | 1.35 |
| adenine nucleotide transport | 2 | -1.19 | 1.19 | 1.41 | 1.1 | 1.28 | 1.45 | -1.24 |
| regulation of NFAT protein import into nucleus | 15 | 1.13 | 1.04 | 1.21 | 1.07 | 1.33 | 1.2 | 1.19 |
| positive regulation of NFAT protein import into nucleus | 15 | 1.13 | 1.04 | 1.21 | 1.07 | 1.33 | 1.2 | 1.19 |
| iron-sulfur cluster binding | 46 | 1.07 | 1.12 | 1.15 | 1.02 | -1.01 | 1.33 | -1.01 |
| 2 iron, 2 sulfur cluster binding | 15 | -1.23 | 1.2 | 1.11 | 1.01 | -1.04 | -1.03 | -1.02 |
| 4 iron, 4 sulfur cluster binding | 15 | 1.46 | 1.1 | 1.27 | -1.03 | 1.06 | 1.96 | 1.05 |
| metal cluster binding | 46 | 1.07 | 1.12 | 1.15 | 1.02 | -1.01 | 1.33 | -1.01 |
| mitochondrial calcium ion homeostasis | 1 | 1.37 | 1.05 | 1.44 | -1.12 | 1.01 | 2.39 | -1.18 |
| histone H3-K9 methylation | 5 | 1.36 | 1.11 | 1.24 | 1.16 | 1.32 | 1.36 | 1.19 |
| histone H3-K4 methylation | 7 | 1.95 | 1.22 | 1.67 | 1.19 | 1.69 | 1.86 | 1.5 |
| regulation of histone H3-K4 methylation | 1 | -1.35 | 1.14 | 1.74 | 1.14 | 1.47 | 1.35 | 1.12 |
| positive regulation of histone H3-K4 methylation | 1 | -1.35 | 1.14 | 1.74 | 1.14 | 1.47 | 1.35 | 1.12 |
| regulation of neurotransmitter transport | 11 | 1.29 | 1.05 | 1.09 | 1.01 | 1.04 | 1.26 | 1.14 |
| response to calcium ion | 3 | -1.58 | -1.06 | -1.06 | 1.03 | 1.08 | -1.26 | -1.33 |
| detection of glucose | 1 | -1.49 | -1.13 | 1.17 | 1.11 | 1.02 | -1.25 | -1.24 |
| response to methylmercury | 1 | -1.7 | -15.86 | -19.15 | -19.05 | -4.94 | 3.57 | 1.25 |
| meiotic recombination checkpoint | 1 | 1.5 | 1.02 | 1.68 | -1.05 | 1.78 | 1.86 | 1.49 |
| protein maturation | 35 | 1.25 | 1.0 | 1.16 | 1.01 | 1.14 | 1.15 | 1.2 |
| detection of stimulus | 142 | 1.02 | -1.06 | -1.01 | 1.02 | -1.0 | 1.02 | -1.09 |
| defense response to virus | 11 | 2.24 | -1.18 | 1.31 | -1.13 | 1.34 | 1.77 | 1.19 |
| histamine transport | 1 | 9.63 | -2.21 | -1.97 | -1.79 | -1.94 | -1.46 | -1.99 |
| serotonin uptake | 1 | -1.06 | -1.11 | -1.0 | -1.08 | -1.15 | 3.37 | -1.04 |
| bacterial cell surface binding | 13 | 1.62 | -1.3 | -1.27 | -1.27 | -1.21 | 1.78 | -1.04 |
| Gram-negative bacterial cell surface binding | 6 | 2.33 | -1.4 | -1.19 | -1.23 | -1.04 | 2.23 | 1.13 |
| Gram-positive bacterial cell surface binding | 1 | 1.05 | 1.01 | 1.06 | 1.12 | 1.02 | -1.02 | -1.17 |
| organelle localization | 87 | 1.05 | 1.18 | 1.31 | 1.13 | 1.34 | 1.21 | 1.16 |
| cellular localization | 545 | 1.05 | 1.13 | 1.26 | 1.13 | 1.25 | 1.15 | 1.12 |
| centrosome localization | 6 | 1.32 | 1.16 | 1.58 | 1.07 | 1.39 | 1.47 | 1.24 |
| endoplasmic reticulum localization | 1 | 1.43 | 1.44 | -1.25 | 1.13 | 1.04 | -1.73 | 1.42 |
| Golgi localization | 1 | 1.07 | 1.7 | 2.32 | 1.13 | 1.82 | 2.48 | 3.32 |
| mitochondrion localization | 9 | -1.42 | 1.39 | 1.56 | 1.16 | 1.41 | 1.21 | 1.26 |
| nucleus localization | 28 | 1.08 | 1.16 | 1.28 | 1.12 | 1.28 | 1.24 | 1.16 |
| vesicle localization | 13 | -1.05 | 1.23 | 1.42 | 1.15 | 1.35 | 1.3 | 1.29 |
| establishment of localization in cell | 458 | 1.03 | 1.13 | 1.26 | 1.13 | 1.25 | 1.13 | 1.1 |
| Yb body | 5 | -1.01 | 1.06 | -1.01 | 1.12 | 1.04 | 1.16 | 1.0 |
| establishment of vesicle localization | 12 | -1.08 | 1.24 | 1.4 | 1.15 | 1.31 | 1.23 | 1.25 |
| maintenance of location in cell | 38 | 1.15 | 1.13 | 1.29 | 1.07 | 1.32 | 1.24 | 1.25 |
| spindle localization | 15 | -1.09 | 1.17 | 1.26 | 1.1 | 1.29 | 1.1 | 1.08 |
| establishment of mitochondrion localization | 5 | -1.72 | 1.29 | 1.54 | 1.18 | 1.39 | 1.16 | 1.27 |
| establishment of organelle localization | 75 | 1.06 | 1.16 | 1.3 | 1.14 | 1.34 | 1.17 | 1.14 |
| maintenance of organelle location | 1 | 1.08 | 1.36 | 1.49 | -1.03 | 1.39 | 1.74 | 1.47 |
| maintenance of nucleus location | 1 | 1.08 | 1.36 | 1.49 | -1.03 | 1.39 | 1.74 | 1.47 |
| oocyte nucleus localization involved in oocyte dorsal/ventral axis specification | 10 | 1.34 | 1.14 | 1.18 | 1.1 | 1.17 | 1.62 | 1.12 |
| nuclear pore localization | 1 | 1.83 | -1.06 | 1.02 | 1.15 | 1.17 | 1.57 | -1.03 |
| localization within membrane | 4 | -1.25 | 1.15 | 1.55 | 1.16 | 1.41 | 1.33 | -1.14 |
| localization of cell | 205 | 1.13 | 1.09 | 1.32 | 1.04 | 1.35 | 1.21 | 1.21 |
| establishment of Golgi localization | 1 | 1.07 | 1.7 | 2.32 | 1.13 | 1.82 | 2.48 | 3.32 |
| establishment of ER localization | 1 | 1.43 | 1.44 | -1.25 | 1.13 | 1.04 | -1.73 | 1.42 |
| cellular oligosaccharide metabolic process | 1 | -1.04 | -1.0 | 1.06 | -1.08 | -1.02 | -1.02 | -1.17 |
| actin filament capping | 2 | -1.38 | -1.03 | 2.18 | 1.01 | 1.59 | -1.39 | -1.04 |
| interaction with host | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| multi-organism process | 285 | 1.15 | -1.16 | -1.07 | -1.15 | -1.05 | 1.17 | 1.13 |
| behavioral interaction between organisms | 120 | 1.07 | -1.05 | 1.06 | -1.03 | 1.01 | 1.21 | 1.15 |
| cellular response to stimulus | 1171 | 1.16 | 1.04 | 1.16 | 1.04 | 1.18 | 1.17 | 1.11 |
| UTP-monosaccharide-1-phosphate uridylyltransferase activity | 1 | 1.07 | -1.0 | -1.14 | -1.11 | -1.05 | -1.01 | 1.08 |
| inositol tetrakisphosphate kinase activity | 1 | -1.15 | 1.56 | -1.07 | 1.07 | -1.15 | -1.25 | 1.09 |
| inositol trisphosphate kinase activity | 5 | 1.28 | 1.12 | 1.04 | -1.27 | 1.01 | 1.27 | 1.33 |
| response to redox state | 1 | 1.13 | 1.09 | 1.52 | 1.04 | 1.43 | -1.06 | -1.03 |
| behavioral response to nutrient | 5 | -1.0 | -1.02 | 1.2 | 1.04 | 1.13 | 1.24 | 1.12 |
| positive regulation of cell division | 1 | -1.14 | 1.41 | 1.3 | 1.24 | 1.17 | 1.85 | 1.43 |
| regulation of nuclear division | 45 | 1.26 | 1.07 | 1.15 | 1.07 | 1.27 | 1.23 | 1.12 |
| negative regulation of nuclear division | 15 | 1.05 | 1.11 | 1.17 | 1.19 | 1.35 | 1.07 | 1.0 |
| positive regulation of nuclear division | 2 | -1.01 | 1.03 | 1.05 | 1.11 | 1.69 | -1.0 | -1.02 |
| response to growth factor stimulus | 13 | 1.49 | -1.07 | 1.91 | -1.01 | 1.18 | 1.43 | 1.11 |
| phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| entry into cell of other organism involved in symbiotic interaction | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| entry into other organism involved in symbiotic interaction | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| histone demethylase activity (H3-K36 specific) | 2 | 1.08 | 1.24 | 1.45 | 1.25 | 1.55 | 1.06 | 1.05 |
| protein autoubiquitination | 1 | 1.42 | -1.23 | 1.7 | 1.22 | 1.08 | 1.57 | -1.21 |
| regulation of mitochondrial membrane potential | 1 | 1.38 | 1.05 | 1.33 | 1.44 | 1.46 | 1.81 | -1.29 |
| regulation of cardioblast differentiation | 8 | 1.02 | 1.09 | 1.19 | 1.04 | 1.21 | -1.01 | 1.39 |
| regulation of focal adhesion assembly | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| negative regulation of focal adhesion assembly | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| regulation of protein kinase B signaling cascade | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| negative regulation of protein kinase B signaling cascade | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| membrane depolarization | 2 | 1.57 | 1.42 | -5.94 | -5.79 | -4.29 | -1.73 | 2.07 |
| peroxiredoxin activity | 6 | -1.25 | 1.02 | -1.13 | 1.12 | 1.03 | -1.35 | -1.22 |
| regulation of calcium ion transport | 9 | 1.35 | 1.01 | 1.15 | 1.0 | 1.06 | 1.49 | 1.16 |
| regulation of calcium ion transport via voltage-gated calcium channel activity | 3 | 1.48 | -1.08 | 1.0 | -1.12 | -1.07 | 1.3 | -1.09 |
| negative regulation of calcium ion transport | 1 | -1.32 | 1.16 | 1.21 | -1.14 | -1.21 | 1.47 | 1.08 |
| positive regulation of calcium ion transport | 2 | 2.64 | -1.06 | 1.57 | 1.01 | 1.45 | 3.25 | 1.94 |
| L-glutamate import | 1 | -51.3 | 1.56 | -15.93 | -1.24 | 2.78 | -3.27 | -134.18 |
| regulation of nervous system development | 135 | 1.05 | 1.12 | 1.33 | 1.08 | 1.3 | 1.18 | 1.16 |
| negative regulation of nervous system development | 13 | -1.19 | 1.12 | 1.25 | 1.16 | 1.16 | -1.1 | 1.05 |
| positive regulation of nervous system development | 8 | -1.2 | 1.13 | 1.29 | 1.1 | 1.32 | 1.04 | 1.35 |
| regulation of synapse assembly | 42 | -1.03 | 1.11 | 1.32 | 1.09 | 1.24 | 1.1 | 1.19 |
| negative regulation of synapse assembly | 13 | -1.19 | 1.12 | 1.25 | 1.16 | 1.16 | -1.1 | 1.05 |
| positive regulation of synapse assembly | 8 | -1.2 | 1.13 | 1.29 | 1.1 | 1.32 | 1.04 | 1.35 |
| regulation of synaptic transmission, glutamatergic | 1 | 1.23 | 1.13 | 1.15 | 1.14 | 1.01 | 1.03 | 1.02 |
| regulation of transmission of nerve impulse | 36 | 1.13 | 1.11 | 1.3 | 1.02 | 1.17 | 1.29 | 1.22 |
| negative regulation of transmission of nerve impulse | 2 | -1.06 | 1.07 | 1.19 | 1.01 | 1.07 | 1.19 | 1.07 |
| positive regulation of transmission of nerve impulse | 2 | -1.81 | 1.29 | 2.08 | -1.04 | 1.39 | 1.52 | 1.29 |
| Se-methyltransferase activity | 2 | -2.66 | 1.46 | -3.05 | 1.35 | 1.97 | -1.55 | -1.14 |
| protein carboxyl O-methyltransferase activity | 2 | 1.17 | 1.1 | 1.15 | 1.29 | 1.38 | 1.07 | -1.31 |
| movement in host environment | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| positive energy taxis | 2 | 1.78 | -1.02 | 1.5 | -1.1 | 1.34 | 2.13 | 1.12 |
| negative energy taxis | 1 | -1.56 | 1.9 | 2.35 | 1.42 | 1.33 | 1.03 | 1.8 |
| movement in environment of other organism involved in symbiotic interaction | 2 | 1.04 | 1.05 | 1.19 | -1.29 | -1.39 | 2.64 | 1.17 |
| regulation of peptidase activity | 20 | 1.05 | 1.09 | 1.25 | 1.18 | 1.38 | 1.06 | 1.03 |
| regulation of endopeptidase activity | 19 | 1.05 | 1.1 | 1.25 | 1.19 | 1.39 | 1.05 | -1.01 |
| imidazole-containing compound metabolic process | 2 | 1.02 | -1.27 | 1.33 | 1.09 | 1.3 | 1.8 | 1.14 |
| inositol monophosphate phosphatase activity | 6 | 1.86 | -1.14 | -1.69 | -1.19 | 1.02 | 1.47 | 1.43 |
| phosphatidylinositol phosphate phosphatase activity | 5 | 1.04 | 1.17 | 1.35 | 1.12 | 1.16 | 1.2 | 1.1 |
| muscle cell development | 82 | 1.48 | 1.06 | 1.26 | -1.03 | 1.2 | 1.47 | 1.18 |
| striated muscle cell development | 81 | 1.48 | 1.06 | 1.26 | -1.03 | 1.2 | 1.48 | 1.18 |
| cardiac cell development | 1 | 1.07 | 1.18 | 1.43 | 1.07 | 1.41 | 1.26 | 1.4 |
| cardiac muscle cell differentiation | 2 | 1.04 | 1.06 | 1.29 | 1.05 | 1.22 | 1.1 | 1.23 |
| cardiac muscle cell development | 1 | 1.07 | 1.18 | 1.43 | 1.07 | 1.41 | 1.26 | 1.4 |
| cortical microtubule | 1 | -1.44 | 1.19 | -1.03 | 1.43 | 1.32 | -1.27 | -1.21 |
| nuclear DNA-directed RNA polymerase complex | 23 | 1.32 | 1.14 | 1.23 | 1.25 | 1.26 | 1.12 | 1.07 |
| Bolwig's organ development | 9 | 1.23 | -1.01 | 1.15 | 1.06 | 1.23 | 1.18 | 1.03 |
| recycling endosome | 4 | -1.11 | 1.29 | 1.32 | 1.11 | 1.24 | 1.01 | 1.18 |
| neuroblast division | 30 | -1.27 | 1.17 | 1.36 | 1.17 | 1.41 | -1.04 | 1.04 |
| asymmetric neuroblast division | 29 | -1.27 | 1.18 | 1.38 | 1.18 | 1.43 | -1.04 | 1.05 |
| asymmetric neuroblast division resulting in ganglion mother cell formation | 2 | -1.54 | 1.67 | 1.31 | 1.18 | 1.85 | -1.54 | 1.65 |
| metal ion homeostasis | 21 | 1.18 | -1.09 | 1.18 | -1.08 | -1.18 | 1.19 | -1.02 |
| divalent inorganic cation homeostasis | 12 | 1.64 | 1.02 | 1.45 | -1.07 | -1.01 | 1.57 | 1.18 |
| monovalent inorganic cation homeostasis | 9 | -1.27 | 1.25 | 1.91 | -1.01 | 1.19 | 1.86 | 1.45 |
| zinc ion homeostasis | 2 | -1.14 | 1.23 | 2.03 | 1.09 | 1.14 | 1.23 | 1.14 |
| copper ion homeostasis | 6 | -1.1 | 1.23 | 1.39 | 1.07 | 1.02 | -1.01 | 1.17 |
| iron ion homeostasis | 8 | 1.49 | -1.19 | -1.16 | -1.17 | -1.18 | 1.48 | 1.09 |
| calcium ion homeostasis | 10 | 1.86 | -1.02 | 1.36 | -1.1 | -1.04 | 1.64 | 1.18 |
| transition metal ion homeostasis | 1 | -2.45 | 1.57 | -1.18 | 1.13 | -1.31 | -2.1 | 1.39 |
| cation homeostasis | 36 | 1.04 | 1.04 | 1.42 | -1.04 | -1.04 | 1.32 | 1.1 |
| cellular chemical homeostasis | 58 | -1.02 | -1.02 | 1.04 | -1.15 | -1.07 | 1.16 | 1.05 |
| transmembrane transport | 398 | -1.34 | -1.02 | -1.21 | -1.11 | -1.13 | 1.0 | -1.15 |
| nucleobase-containing small molecule metabolic process | 189 | 1.1 | 1.04 | 1.03 | 1.02 | 1.07 | 1.12 | -1.0 |
| lipid homeostasis | 17 | -1.32 | -1.08 | -1.1 | 1.01 | -1.02 | -1.16 | -1.17 |
| fatty acid homeostasis | 1 | 1.18 | 1.17 | 1.43 | 1.18 | 1.36 | -1.03 | 1.42 |
| acylglycerol homeostasis | 1 | 1.16 | 1.22 | 2.13 | 1.14 | 1.8 | 2.14 | 1.55 |
| phospholipid homeostasis | 1 | -1.95 | 1.39 | 1.68 | 1.01 | 1.49 | -1.24 | 1.08 |
| sterol homeostasis | 4 | -4.57 | -1.96 | -1.16 | 1.4 | -1.7 | -2.27 | -3.58 |
| response to hyperoxia | 1 | 1.14 | 1.04 | 1.05 | 1.06 | 1.07 | 1.1 | 1.08 |
| entry into diapause | 1 | -1.14 | 1.2 | 1.7 | 1.09 | 2.04 | 1.39 | 1.74 |
| digestive system development | 87 | -1.01 | -1.06 | 1.09 | -1.05 | 1.19 | 1.11 | 1.14 |
| reflex | 5 | -1.0 | -1.02 | 1.2 | 1.04 | 1.13 | 1.24 | 1.12 |
| rhythmic synaptic transmission | 1 | 9.13 | 1.03 | 1.44 | -1.07 | -1.11 | 1.07 | 1.06 |
| regulation of synaptic activity | 5 | 1.07 | 1.03 | 1.09 | -1.04 | 1.05 | 1.27 | 1.1 |
| convergent extension | 2 | 1.27 | 1.22 | 1.43 | 1.14 | 1.36 | 1.4 | 1.35 |
| convergent extension involved in gastrulation | 2 | 1.27 | 1.22 | 1.43 | 1.14 | 1.36 | 1.4 | 1.35 |
| mediolateral intercalation | 2 | 1.27 | 1.22 | 1.43 | 1.14 | 1.36 | 1.4 | 1.35 |
| retina development in camera-type eye | 1 | -1.1 | -1.15 | -1.07 | -1.18 | 1.52 | 1.28 | 1.24 |
| heart contraction | 3 | -1.25 | 1.41 | -1.37 | -1.01 | 1.12 | 1.05 | 1.18 |
| cardiac muscle contraction | 1 | -2.87 | 2.31 | -2.71 | -1.16 | 1.45 | 1.26 | 1.68 |
| regulation of protein glycosylation | 1 | -1.37 | -1.13 | -1.19 | -1.54 | 1.82 | 7.35 | 2.8 |
| neurofilament cytoskeleton organization | 1 | 1.1 | 1.12 | 1.07 | -1.03 | -1.01 | 1.1 | 1.01 |
| canonical Wnt receptor signaling pathway | 8 | 1.24 | 1.0 | 1.39 | 1.04 | 1.59 | 1.21 | 1.49 |
| Wnt receptor signaling pathway, planar cell polarity pathway | 4 | 1.06 | -1.1 | 1.26 | -1.16 | 1.25 | 1.67 | 1.42 |
| large conductance calcium-activated potassium channel activity | 1 | -2.53 | -3.67 | 1.72 | 2.84 | -2.74 | -4.98 | -3.6 |
| synapse maturation | 2 | -1.1 | 1.15 | 1.09 | 1.05 | 1.27 | 1.2 | 1.3 |
| regulation of postsynaptic membrane potential | 4 | 1.21 | 1.34 | -2.02 | -2.31 | -1.7 | 1.03 | 1.76 |
| regulation of excitatory postsynaptic membrane potential | 2 | 1.57 | 1.42 | -5.94 | -5.79 | -4.29 | -1.73 | 2.07 |
| molecular transducer activity | 443 | 1.09 | 1.01 | 1.1 | 1.0 | 1.08 | 1.12 | 1.06 |
| binding, bridging | 17 | -1.01 | 1.02 | 1.14 | 1.03 | 1.09 | 1.16 | 1.1 |
| limb development | 54 | -1.01 | 1.08 | 1.1 | 1.01 | 1.36 | 1.25 | 1.25 |
| male mating behavior | 44 | -1.08 | -1.14 | 1.03 | -1.05 | -1.05 | -1.07 | 1.07 |
| female mating behavior | 24 | -1.01 | -1.19 | -1.15 | -1.08 | -1.06 | 1.22 | 1.13 |
| cell pole | 3 | 1.46 | 1.08 | 1.28 | 1.11 | 1.49 | 1.29 | 1.1 |
| regulation of lipase activity | 8 | 1.04 | 1.1 | 1.09 | 1.07 | 1.01 | 1.07 | 1.0 |
| negative regulation of lipase activity | 2 | -1.21 | 1.05 | -1.16 | 1.19 | -1.24 | -1.13 | -1.15 |
| positive regulation of lipase activity | 6 | 1.13 | 1.12 | 1.17 | 1.04 | 1.09 | 1.14 | 1.05 |
| regulation of nuclear-transcribed mRNA poly(A) tail shortening | 4 | 1.18 | 1.11 | 1.21 | 1.11 | 1.04 | 1.21 | 1.05 |
| positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 4 | 1.18 | 1.11 | 1.21 | 1.11 | 1.04 | 1.21 | 1.05 |
| regulation of retinal cone cell fate commitment | 2 | -1.43 | 1.42 | 1.17 | 1.08 | 1.59 | -1.55 | 1.31 |
| lipase activator activity | 2 | -1.41 | 1.32 | 1.65 | 1.18 | 1.23 | 1.2 | -1.03 |
| delamination | 8 | 1.85 | 1.18 | 1.27 | 1.06 | 1.22 | 1.87 | 1.48 |
| oenocyte delamination | 2 | 2.23 | 1.23 | 1.34 | 1.08 | 1.47 | 1.18 | 1.41 |
| regulation of mitotic spindle organization | 5 | -1.03 | 1.11 | 1.19 | 1.15 | 1.35 | 1.05 | -1.07 |
| anatomical structure homeostasis | 48 | 1.3 | 1.03 | 1.25 | 1.01 | 1.27 | 1.36 | 1.27 |
| germ-line stem-cell niche homeostasis | 4 | 1.24 | 1.27 | 1.46 | 1.22 | 1.66 | 1.51 | 1.42 |
| regulation of glial cell proliferation | 4 | 1.83 | 1.22 | 1.55 | -1.02 | 1.23 | 1.76 | 1.45 |
| positive regulation of glial cell proliferation | 2 | 2.9 | -1.05 | 1.75 | -1.19 | 1.16 | 1.75 | 1.33 |
| negative regulation of glial cell proliferation | 2 | 1.15 | 1.57 | 1.36 | 1.13 | 1.31 | 1.77 | 1.58 |
| regulation of macromolecule metabolic process | 998 | 1.18 | 1.09 | 1.19 | 1.07 | 1.19 | 1.2 | 1.13 |
| regulation of transcription initiation from RNA polymerase II promoter | 5 | 1.43 | 1.28 | 1.45 | 1.26 | 1.36 | 1.28 | 1.29 |
| positive regulation of transcription initiation from RNA polymerase II promoter | 2 | 1.1 | 1.32 | 1.52 | 1.2 | 1.24 | 1.14 | 1.45 |
| cilium morphogenesis | 33 | -1.14 | 1.06 | 1.0 | 1.05 | -1.01 | -1.08 | -1.05 |
| regulation of ovulation | 2 | 1.79 | 1.12 | 1.06 | 1.1 | 1.24 | 1.64 | 1.35 |
| regulation of oocyte development | 25 | -1.02 | 1.16 | 1.34 | 1.11 | 1.28 | 1.31 | 1.17 |
| regulation of cell development | 174 | 1.05 | 1.11 | 1.34 | 1.08 | 1.3 | 1.21 | 1.18 |
| formation of a compartment boundary | 1 | 1.36 | -1.06 | 1.89 | 1.07 | 1.62 | -1.06 | 1.23 |
| long term synaptic depression | 1 | -1.13 | 1.08 | 1.37 | 1.02 | 1.39 | 1.34 | 1.23 |
| germ plasm | 13 | 1.42 | 1.03 | 1.1 | 1.02 | 1.09 | 1.51 | -1.01 |
| regulation of sarcomere organization | 1 | 6.86 | -1.53 | -1.01 | -1.17 | 1.13 | 2.97 | 1.11 |
| regulation of cell diameter | 3 | 1.2 | 1.28 | 1.27 | 1.02 | 1.3 | 1.13 | 1.19 |
| GTP cyclohydrolase I regulator activity | 1 | 33.5 | -5.63 | -5.79 | -6.03 | -4.58 | -1.32 | 6.94 |
| head development | 23 | 1.18 | -1.03 | 1.07 | 1.05 | -1.03 | -1.01 | 1.01 |
| cell chemotaxis | 7 | 1.41 | -1.02 | 1.42 | 1.1 | 1.42 | -1.11 | -1.12 |
| regulation of cellular localization | 84 | 1.07 | 1.11 | 1.24 | 1.08 | 1.19 | 1.26 | 1.16 |
| leucine import | 3 | 1.2 | -1.03 | -1.17 | 1.06 | 1.15 | -1.13 | 2.49 |
| flight | 4 | 2.08 | 1.01 | -1.1 | -1.13 | 1.05 | 2.84 | 1.39 |
| innervation | 1 | -2.0 | 1.87 | 1.43 | 1.14 | 2.42 | -2.25 | 1.78 |
| axonogenesis involved in innervation | 1 | -2.0 | 1.87 | 1.43 | 1.14 | 2.42 | -2.25 | 1.78 |
| vitelline envelope | 4 | -1.15 | 1.03 | -1.1 | -1.06 | -1.17 | -1.2 | -1.07 |
| cytosolic calcium ion transport | 1 | 2.52 | 1.29 | 3.07 | -1.16 | 1.37 | 5.13 | 2.97 |
| calcium ion transport into cytosol | 1 | 2.52 | 1.29 | 3.07 | -1.16 | 1.37 | 5.13 | 2.97 |
| epithelium development | 247 | 1.17 | 1.05 | 1.28 | 1.02 | 1.3 | 1.33 | 1.27 |
| trachea development | 3 | 1.59 | -1.2 | 1.46 | -1.32 | -1.06 | 1.27 | -1.04 |
| trachea morphogenesis | 1 | 4.21 | -1.18 | -1.09 | -1.07 | -1.3 | -1.11 | -1.25 |
| branching involved in open tracheal system development | 45 | 1.12 | 1.12 | 1.47 | 1.03 | 1.34 | 1.29 | 1.3 |
| mesenchyme development | 1 | 1.47 | 1.13 | -1.02 | 1.1 | -1.07 | -1.03 | 1.1 |
| regulation of cell projection assembly | 14 | 1.07 | 1.12 | 1.53 | 1.12 | 1.42 | -1.16 | 1.1 |
| muscle tissue development | 86 | 1.17 | 1.09 | 1.24 | 1.03 | 1.18 | 1.33 | 1.16 |
| skeletal muscle organ development | 104 | 1.28 | 1.09 | 1.25 | 1.02 | 1.22 | 1.35 | 1.19 |
| respiratory system development | 181 | 1.4 | 1.03 | 1.27 | -1.03 | 1.25 | 1.52 | 1.28 |
| negative regulation of cell death | 56 | 1.23 | 1.09 | 1.31 | 1.02 | 1.24 | 1.34 | 1.27 |
| developmental growth involved in morphogenesis | 50 | 1.3 | 1.12 | 1.32 | 1.02 | 1.35 | 1.3 | 1.31 |
| epithelial tube morphogenesis | 85 | 1.18 | 1.0 | 1.24 | -1.0 | 1.3 | 1.38 | 1.17 |
| negative regulation of mitotic anaphase-promoting complex activity | 1 | 1.67 | 1.26 | 1.32 | 1.4 | 1.23 | 1.13 | 1.21 |
| regulation of peptide hormone processing | 1 | -2.9 | -1.14 | 1.64 | 1.56 | 2.18 | -4.01 | -1.31 |
| negative regulation of peptide hormone processing | 1 | -2.9 | -1.14 | 1.64 | 1.56 | 2.18 | -4.01 | -1.31 |
| morphogenesis of an epithelial fold | 7 | 1.32 | 1.05 | 1.42 | 1.01 | 1.41 | 1.46 | 1.64 |
| morphogenesis of an epithelial bud | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| multicellular organismal iron ion homeostasis | 1 | -2.45 | 1.57 | -1.18 | 1.13 | -1.31 | -2.1 | 1.39 |
| nucleoside-triphosphatase regulator activity | 126 | 1.07 | 1.14 | 1.34 | 1.04 | 1.26 | 1.22 | 1.19 |
| ATPase regulator activity | 4 | 1.49 | 1.1 | 1.13 | 1.22 | 1.27 | 1.15 | 1.16 |
| regulation of vesicle-mediated transport | 25 | 1.05 | 1.04 | 1.38 | 1.1 | 1.22 | 1.61 | 1.14 |
| regulation of ER to Golgi vesicle-mediated transport | 1 | -1.04 | 1.0 | 1.09 | 1.3 | 1.23 | -1.23 | -1.03 |
| regulation of microtubule-based movement | 2 | 1.32 | 1.15 | 1.52 | 1.06 | 1.31 | 2.78 | 1.59 |
| regulation of endoribonuclease activity | 1 | 1.58 | 1.16 | -1.09 | 1.12 | 1.04 | 1.36 | -1.13 |
| regulation of ribonuclease activity | 1 | 1.58 | 1.16 | -1.09 | 1.12 | 1.04 | 1.36 | -1.13 |
| negative regulation of ribonuclease activity | 1 | 1.58 | 1.16 | -1.09 | 1.12 | 1.04 | 1.36 | -1.13 |
| negative regulation of endoribonuclease activity | 1 | 1.58 | 1.16 | -1.09 | 1.12 | 1.04 | 1.36 | -1.13 |
| ectodermal placode formation | 4 | -1.14 | 1.1 | 1.23 | 1.12 | 1.47 | -1.2 | -1.08 |
| cell fate commitment involved in formation of primary germ layers | 17 | 1.6 | 1.01 | 1.08 | -1.06 | 1.04 | 1.29 | 1.13 |
| intracellular mRNA localization involved in pattern specification process | 42 | 1.09 | 1.16 | 1.32 | 1.13 | 1.25 | 1.24 | 1.19 |
| intracellular mRNA localization involved in anterior/posterior axis specification | 41 | 1.07 | 1.17 | 1.33 | 1.14 | 1.26 | 1.24 | 1.19 |
| establishment of glial blood-brain barrier | 16 | 2.59 | -1.2 | 1.31 | -1.17 | 1.29 | 2.35 | 1.44 |
| cardiac cell fate commitment | 9 | 1.12 | 1.05 | 1.08 | 1.02 | 1.08 | 1.04 | 1.06 |
| cardiac cell fate specification | 1 | -1.42 | 1.01 | 1.2 | 1.29 | 2.44 | -1.59 | 1.35 |
| cardiac cell fate determination | 4 | 1.32 | 1.0 | 1.08 | -1.04 | -1.05 | -1.0 | 1.04 |
| heart formation | 6 | 1.11 | -1.07 | 1.07 | 1.07 | 1.2 | 1.27 | 1.2 |
| regulation of gene silencing by RNA | 8 | 1.82 | 1.17 | 1.15 | 1.09 | 1.12 | 1.83 | 1.43 |
| negative regulation of gene silencing by RNA | 5 | 1.71 | 1.18 | 1.16 | 1.09 | 1.12 | 2.11 | 1.55 |
| regulation of gene silencing | 34 | 1.39 | 1.19 | 1.17 | 1.11 | 1.2 | 1.51 | 1.4 |
| negative regulation of gene silencing | 12 | 1.32 | 1.18 | 1.16 | 1.15 | 1.16 | 1.51 | 1.32 |
| left/right pattern formation | 1 | -1.92 | 1.09 | 4.17 | -1.09 | 2.38 | -1.63 | 1.77 |
| kidney morphogenesis | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| regulation of mRNA catabolic process | 4 | 1.18 | 1.11 | 1.21 | 1.11 | 1.04 | 1.21 | 1.05 |
| positive regulation of mRNA catabolic process | 4 | 1.18 | 1.11 | 1.21 | 1.11 | 1.04 | 1.21 | 1.05 |
| membrane organization | 261 | 1.07 | 1.1 | 1.21 | 1.08 | 1.23 | 1.2 | 1.13 |
| membrane fusion | 17 | -1.08 | 1.06 | 1.29 | 1.18 | 1.2 | 1.18 | -1.03 |
| endodermal digestive tract morphogenesis | 1 | -5.18 | 1.12 | -1.52 | 1.19 | -1.17 | -3.69 | 1.24 |
| ovum-producing ovary development | 1 | -1.49 | 2.58 | -1.26 | -1.23 | -1.23 | -1.38 | 1.24 |
| regulation of wound healing | 1 | 9.2 | -1.31 | -1.17 | -1.11 | -1.07 | 7.07 | 1.71 |
| peptidoglycan recognition protein signaling pathway | 8 | 1.83 | 1.08 | 1.26 | 1.07 | 1.21 | 1.54 | 1.2 |
| regulation of peptidoglycan recognition protein signaling pathway | 4 | 2.19 | -1.16 | 1.49 | 1.19 | -1.07 | -1.21 | -1.53 |
| positive regulation of peptidoglycan recognition protein signaling pathway | 1 | 8.35 | -1.25 | -1.3 | -1.07 | -1.15 | 1.62 | -1.07 |
| negative regulation of peptidoglycan recognition protein signaling pathway | 3 | 1.4 | -1.13 | 1.86 | 1.28 | -1.04 | -1.52 | -1.72 |
| muscle structure development | 204 | 1.33 | 1.03 | 1.2 | 1.01 | 1.19 | 1.28 | 1.19 |
| regulation of neural retina development | 2 | -1.43 | 1.42 | 1.17 | 1.08 | 1.59 | -1.55 | 1.31 |
| chaperone-mediated protein folding | 4 | 1.23 | 1.17 | 1.41 | 1.19 | 1.43 | -1.13 | 1.19 |
| regulation of protein tyrosine kinase activity | 5 | 1.47 | 1.14 | 1.46 | -1.01 | 1.32 | 1.52 | 1.27 |
| positive regulation of protein tyrosine kinase activity | 1 | -1.21 | 1.16 | 2.16 | -1.11 | 1.4 | 1.68 | 1.37 |
| negative regulation of protein tyrosine kinase activity | 4 | 1.69 | 1.13 | 1.32 | 1.01 | 1.3 | 1.49 | 1.25 |
| peptidase regulator activity | 83 | 1.36 | -1.46 | -1.4 | -1.48 | -1.27 | 1.43 | 1.07 |
| morphogenesis of a branching epithelium | 45 | 1.12 | 1.12 | 1.47 | 1.03 | 1.34 | 1.29 | 1.3 |
| establishment of monopolar cell polarity | 10 | 1.54 | 1.16 | 1.19 | -1.02 | 1.43 | 1.47 | 1.4 |
| type I terminal button | 2 | 2.85 | 1.08 | 1.12 | -1.03 | -1.01 | 2.59 | 1.3 |
| establishment or maintenance of bipolar cell polarity | 32 | 1.32 | 1.09 | 1.43 | 1.03 | 1.37 | 1.42 | 1.25 |
| cell surface receptor signaling pathway involved in heart development | 3 | 1.09 | 1.1 | 1.13 | 1.08 | 1.18 | 1.14 | 2.09 |
| canonical Wnt receptor signaling pathway involved in heart development | 3 | 1.09 | 1.1 | 1.13 | 1.08 | 1.18 | 1.14 | 2.09 |
| renal filtration cell differentiation | 11 | 1.64 | -1.04 | 1.33 | -1.09 | 1.12 | 1.26 | 1.37 |
| nephrocyte differentiation | 11 | 1.64 | -1.04 | 1.33 | -1.09 | 1.12 | 1.26 | 1.37 |
| pericardial nephrocyte differentiation | 9 | 1.9 | -1.04 | 1.48 | -1.11 | 1.17 | 1.39 | 1.52 |
| garland nephrocyte differentiation | 2 | -1.2 | -1.04 | -1.19 | 1.02 | -1.08 | -1.21 | -1.15 |
| renal tubule development | 62 | 1.15 | -1.03 | 1.11 | -1.01 | 1.22 | 1.34 | 1.08 |
| anterior Malpighian tubule development | 12 | -1.05 | -1.01 | 1.18 | -1.15 | 1.13 | 1.62 | 1.06 |
| posterior Malpighian tubule development | 2 | 1.35 | -1.06 | 1.02 | -1.25 | -1.0 | 1.06 | 2.86 |
| Malpighian tubule stellate cell differentiation | 3 | 1.38 | 1.01 | 1.13 | 1.37 | -1.02 | -1.07 | -1.11 |
| epithelial cell proliferation involved in Malpighian tubule morphogenesis | 11 | 1.14 | 1.03 | 1.13 | -1.0 | 1.25 | 1.04 | 1.44 |
| Malpighian tubule bud morphogenesis | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| renal tubule morphogenesis | 44 | 1.29 | -1.1 | 1.07 | -1.04 | 1.27 | 1.38 | 1.09 |
| cell rearrangement involved in Malpighian tubule morphogenesis | 3 | 2.04 | 1.05 | 1.89 | -1.07 | 1.16 | 1.0 | 1.3 |
| cell morphogenesis involved in Malpighian tubule morphogenesis | 1 | 3.86 | 1.45 | 2.61 | -1.03 | 2.28 | 4.0 | 3.57 |
| establishment or maintenance of monopolar cell polarity | 10 | 1.54 | 1.16 | 1.19 | -1.02 | 1.43 | 1.47 | 1.4 |
| neural precursor cell proliferation | 40 | -1.13 | 1.17 | 1.36 | 1.16 | 1.4 | 1.07 | 1.11 |
| cell chemotaxis involved in Malpighian tubule morphogenesis | 3 | 2.04 | 1.05 | 1.89 | -1.07 | 1.16 | 1.0 | 1.3 |
| BMP signaling pathway involved in Malpighian tubule cell chemotaxis | 3 | 2.04 | 1.05 | 1.89 | -1.07 | 1.16 | 1.0 | 1.3 |
| regulation of Wnt protein secretion | 3 | 1.21 | 1.05 | 1.22 | 1.06 | -1.1 | -1.07 | 1.29 |
| positive regulation of Wnt protein secretion | 3 | 1.21 | 1.05 | 1.22 | 1.06 | -1.1 | -1.07 | 1.29 |
| Malpighian tubule tip cell differentiation | 9 | -1.2 | 1.13 | 1.24 | 1.07 | 1.14 | -1.23 | 1.24 |
| regulation of extent of cell growth | 7 | -1.17 | 1.19 | 1.37 | 1.14 | 1.29 | 1.18 | 1.05 |
| specification of axis polarity | 5 | 1.18 | 1.04 | 1.06 | 1.17 | 1.12 | -1.03 | 1.06 |
| intracellular protein transmembrane transport | 5 | -1.62 | 1.28 | -1.17 | -1.01 | -1.28 | -1.9 | 1.13 |
| macromolecular complex assembly | 152 | 1.06 | 1.1 | 1.27 | 1.14 | 1.27 | 1.1 | 1.02 |
| protein-DNA complex assembly | 42 | 1.03 | 1.08 | 1.27 | 1.16 | 1.28 | -1.05 | 1.02 |
| biological regulation | 2649 | 1.15 | 1.05 | 1.17 | 1.04 | 1.17 | 1.17 | 1.12 |
| regulation of biological quality | 662 | 1.12 | 1.03 | 1.16 | 1.01 | 1.17 | 1.2 | 1.14 |
| regulation of molecular function | 176 | 1.23 | 1.12 | 1.28 | 1.09 | 1.25 | 1.26 | 1.18 |
| aspartic-type peptidase activity | 19 | -1.6 | -1.03 | 1.15 | -1.16 | -1.46 | -1.86 | -1.32 |
| threonine-type peptidase activity | 23 | 1.34 | -1.02 | 1.1 | 1.11 | 1.12 | -1.15 | -1.1 |
| serine-type exopeptidase activity | 13 | -1.19 | -1.2 | -1.0 | -1.01 | -1.57 | -1.53 | 1.1 |
| intracellular organelle lumen | 588 | 1.2 | 1.12 | 1.23 | 1.16 | 1.24 | 1.22 | -1.0 |
| carbon monoxide binding | 1 | -1.13 | -1.1 | -1.21 | -1.08 | -1.27 | -1.0 | -1.19 |
| nitric oxide binding | 1 | -1.13 | -1.1 | -1.21 | -1.08 | -1.27 | -1.0 | -1.19 |
| purine NTP-dependent helicase activity | 76 | 1.18 | 1.21 | 1.34 | 1.17 | 1.3 | 1.31 | 1.19 |
| neuron homeostasis | 3 | -1.16 | 1.18 | 1.11 | 1.05 | 1.04 | -1.04 | -1.02 |
| apoptosis in response to endoplasmic reticulum stress | 1 | -1.56 | 1.12 | -1.6 | -1.09 | -1.49 | -1.56 | -1.26 |
| proline-rich region binding | 1 | -2.96 | 1.13 | 1.63 | 1.21 | 1.47 | -2.66 | -1.29 |
| proton-transporting two-sector ATPase complex assembly | 2 | 1.21 | 1.11 | 1.39 | 1.2 | 1.21 | 1.72 | -1.33 |
| clustering of voltage-gated calcium channels | 1 | 1.29 | -1.71 | -1.84 | -1.91 | -1.32 | 16.38 | -1.61 |
| histone lysine demethylation | 6 | 1.15 | 1.33 | 1.71 | 1.16 | 1.56 | 1.61 | 1.38 |
| glycosylation | 53 | 1.14 | 1.04 | 1.06 | 1.04 | 1.1 | 1.12 | 1.2 |
| synaptic vesicle budding | 14 | 1.04 | 1.17 | 1.47 | 1.12 | 1.28 | 1.36 | 1.24 |
| occluding junction | 26 | 2.55 | -1.23 | 1.25 | -1.28 | 1.23 | 3.22 | 1.62 |
| anchoring junction | 59 | 1.24 | 1.15 | 1.4 | 1.04 | 1.4 | 1.33 | 1.31 |
| kynurenine metabolic process | 1 | 1.33 | -1.46 | -1.36 | -1.56 | -1.39 | 2.84 | -1.33 |
| chromosome organization involved in meiosis | 6 | 1.05 | 1.07 | 1.09 | 1.05 | 1.18 | -1.08 | -1.05 |
| synaptonemal complex organization | 3 | 1.06 | 1.0 | 1.13 | 1.06 | 1.06 | 1.25 | 1.03 |
| synaptonemal complex disassembly | 1 | 1.38 | -1.04 | 1.41 | 1.16 | 1.34 | 2.25 | 1.03 |
| regulation of establishment of protein localization | 42 | 1.15 | 1.11 | 1.24 | 1.09 | 1.2 | 1.21 | 1.21 |
| actin-mediated cell contraction | 4 | -1.26 | 1.28 | 1.59 | 1.06 | 1.39 | 1.15 | 1.23 |
| tyrosyl-DNA phosphodiesterase activity | 1 | -1.61 | -1.07 | -1.0 | -1.17 | -1.28 | -1.58 | -1.34 |
| protein complex biogenesis | 81 | 1.0 | 1.09 | 1.27 | 1.1 | 1.24 | 1.13 | -1.01 |
| proton-transporting ATP synthase complex biogenesis | 2 | 1.21 | 1.11 | 1.39 | 1.2 | 1.21 | 1.72 | -1.33 |
| phosphatidylinositol-4-phosphate binding | 1 | -2.39 | 1.47 | 1.51 | 1.07 | -1.25 | -2.16 | 1.24 |
| extracellular matrix constituent secretion | 2 | 1.62 | -1.11 | 1.06 | -1.19 | 1.17 | -1.33 | -1.09 |
| radical SAM enzyme activity | 1 | 1.58 | 1.18 | 1.95 | 1.1 | 1.45 | 3.43 | -1.02 |
| ferritin complex | 3 | -1.78 | 1.04 | 1.19 | 1.02 | -1.06 | -2.25 | -1.21 |
| phosphatidic acid binding | 1 | 1.31 | 1.26 | 1.47 | 1.37 | 1.45 | 1.27 | 1.07 |
| cellular response to hydrogen peroxide | 5 | -1.31 | 1.04 | -1.13 | 1.14 | 1.05 | -1.39 | -1.21 |
| regulation of stress-activated protein kinase signaling cascade | 33 | -1.0 | 1.2 | 1.31 | 1.13 | 1.3 | 1.36 | 1.17 |
| negative regulation of stress-activated protein kinase signaling cascade | 12 | 1.09 | 1.13 | 1.44 | 1.11 | 1.23 | 1.18 | 1.08 |
| positive regulation of stress-activated protein kinase signaling cascade | 12 | 1.07 | 1.28 | 1.47 | 1.07 | 1.29 | 1.52 | 1.25 |
| lipoprotein particle receptor binding | 1 | 3.66 | -1.09 | 1.54 | -1.1 | 1.65 | 2.15 | 1.33 |
| triglyceride homeostasis | 10 | -1.54 | -1.28 | -1.3 | -1.03 | -1.36 | -1.47 | -1.44 |
| regulation of ERK1 and ERK2 cascade | 3 | 1.06 | 1.01 | -1.21 | -1.22 | -1.32 | -1.26 | 1.87 |
| positive regulation of ERK1 and ERK2 cascade | 3 | 1.06 | 1.01 | -1.21 | -1.22 | -1.32 | -1.26 | 1.87 |
| high mobility group box 1 binding | 1 | 3.82 | -1.26 | 1.16 | -1.51 | 1.23 | 1.8 | 1.55 |
| chaperone cofactor-dependent protein refolding | 1 | 1.92 | 1.17 | 1.75 | 1.1 | 1.32 | 1.14 | 1.27 |
| NAD+ binding | 7 | 1.31 | 1.11 | -1.1 | 1.29 | -1.04 | 1.13 | 1.25 |
| glutamine binding | 1 | -1.12 | 1.14 | 1.63 | 1.21 | 1.33 | 1.0 | 1.05 |
| carbamoyl phosphate metabolic process | 1 | -1.12 | 1.14 | 1.63 | 1.21 | 1.33 | 1.0 | 1.05 |
| carbamoyl phosphate biosynthetic process | 1 | -1.12 | 1.14 | 1.63 | 1.21 | 1.33 | 1.0 | 1.05 |
| cellular response to cold | 2 | 1.0 | 1.06 | -1.05 | 1.0 | -1.03 | -1.07 | -1.17 |
| nonhomologous end joining complex | 2 | -1.03 | 1.29 | 1.58 | 1.29 | 1.51 | -1.21 | 1.27 |
| elongin complex | 1 | -1.03 | 1.24 | 1.37 | 1.02 | 1.36 | -1.06 | 1.29 |
| cell hair | 2 | 1.46 | 1.0 | 1.38 | -1.02 | 1.32 | 1.45 | 1.21 |
| SAGA-type complex | 20 | 1.38 | 1.1 | 1.14 | 1.26 | 1.22 | 1.03 | -1.02 |
| plus-end specific microtubule depolymerization | 1 | 1.23 | 1.19 | 1.22 | 1.26 | 1.69 | -1.05 | 1.08 |
| respiratory chain | 68 | 1.11 | 1.0 | 1.19 | 1.01 | 1.07 | 1.57 | -1.31 |
| response to oxygen levels | 48 | 1.13 | 1.14 | 1.22 | 1.09 | 1.26 | 1.28 | 1.19 |
| repressing transcription factor binding | 13 | 1.29 | -1.04 | -1.01 | -1.14 | 1.25 | 1.49 | 1.64 |
| regulation of microtubule cytoskeleton organization | 27 | 1.18 | 1.14 | 1.4 | 1.06 | 1.41 | 1.45 | 1.25 |
| death domain binding | 1 | 1.48 | 1.27 | 1.15 | 1.13 | 1.09 | -1.04 | 1.31 |
| histone H3-K36 demethylation | 2 | 1.08 | 1.24 | 1.45 | 1.25 | 1.55 | 1.06 | 1.05 |
| L-tyrosine aminotransferase activity | 1 | 1.14 | 1.12 | -1.09 | -1.15 | 1.09 | -1.15 | -1.05 |
| L-glutamine aminotransferase activity | 2 | -1.0 | -1.44 | 1.48 | -1.44 | 1.38 | 1.11 | 2.78 |
| adenylyltransferase activity | 11 | -1.05 | 1.19 | 1.21 | 1.12 | 1.08 | -1.03 | 1.08 |
| cytidylyltransferase activity | 5 | 1.44 | -1.22 | 1.18 | -1.16 | 1.33 | -1.05 | 1.53 |
| guanylyltransferase activity | 4 | -1.18 | 1.19 | 1.23 | 1.25 | 1.19 | -1.14 | 1.16 |
| uridylyltransferase activity | 2 | -1.21 | -1.06 | 1.2 | -1.1 | 1.09 | -1.36 | 1.36 |
| cadmium ion transmembrane transport | 1 | -1.37 | 1.35 | 1.68 | 1.28 | 1.12 | -1.35 | -1.15 |
| mitochondrion morphogenesis | 2 | 1.03 | 1.2 | 1.34 | 1.27 | 1.61 | 1.2 | -1.12 |
| protein localization in mitochondrion | 28 | 1.07 | 1.09 | 1.21 | 1.13 | 1.12 | 1.18 | -1.1 |
| calcium ion transmembrane transport | 7 | 1.59 | 1.03 | 1.19 | -1.08 | -1.01 | 1.55 | 1.32 |
| dendrite self-avoidance | 7 | 1.17 | 1.14 | 1.29 | 1.13 | 1.33 | 1.54 | 1.21 |
| juvenile hormone response element binding | 2 | -1.22 | 1.25 | 1.18 | 1.1 | 1.78 | -1.07 | 1.14 |
| SWI/SNF-type complex | 10 | 1.12 | 1.16 | 1.37 | 1.17 | 1.42 | 1.35 | 1.27 |
| PBAF complex | 1 | -1.21 | 1.11 | -1.01 | 1.16 | 1.01 | -1.02 | 1.11 |
| regulation of protein processing | 1 | -2.9 | -1.14 | 1.64 | 1.56 | 2.18 | -4.01 | -1.31 |
| nucleosome-dependent ATPase activity | 2 | 1.37 | 1.25 | 2.22 | 1.17 | 1.97 | 1.84 | 1.81 |
| proteasome binding | 1 | 1.49 | 1.1 | 1.49 | 1.27 | 1.56 | -1.75 | 1.05 |
| protein modification by small protein removal | 15 | 1.19 | 1.08 | 1.36 | 1.13 | 1.29 | 1.19 | 1.15 |
| protein modification by small protein conjugation or removal | 62 | 1.15 | 1.12 | 1.38 | 1.15 | 1.28 | 1.17 | 1.13 |
| poly-purine tract binding | 5 | 1.32 | 1.19 | 1.41 | 1.19 | 1.38 | 1.34 | 1.35 |
| cellular macromolecule localization | 266 | 1.06 | 1.17 | 1.3 | 1.13 | 1.24 | 1.16 | 1.17 |
| cyclic nucleotide transport | 1 | 1.9 | -1.14 | -1.37 | 1.0 | -1.16 | 1.35 | -1.12 |
| cGMP transport | 1 | 1.9 | -1.14 | -1.37 | 1.0 | -1.16 | 1.35 | -1.12 |
| spindle envelope | 4 | 1.54 | 1.11 | 1.47 | 1.01 | 1.35 | 1.6 | 1.17 |
| histone H3-K27 methylation | 3 | 1.06 | 1.17 | 1.29 | 1.08 | 1.13 | -1.1 | -1.05 |
| C2H2 zinc finger domain binding | 1 | 1.47 | -1.07 | -1.23 | 1.4 | -1.01 | -1.35 | -1.31 |
| peptidyl-lysine 5-dioxygenase activity | 2 | 1.13 | 1.08 | 1.05 | -1.04 | 1.19 | 1.95 | 1.17 |
| phosphorylation of RNA polymerase II C-terminal domain | 1 | -1.89 | -1.04 | 1.09 | 1.37 | 2.39 | -1.87 | -1.49 |
| protoporphyrinogen oxidase activity | 2 | -1.11 | 1.24 | 1.36 | 1.38 | 1.3 | 1.17 | -1.16 |
| Sin3-type complex | 10 | 1.24 | 1.21 | 1.36 | 1.21 | 1.38 | 1.37 | 1.15 |
| chromatin maintenance | 1 | -1.2 | 1.03 | 1.07 | -1.11 | 1.52 | -1.28 | -1.06 |
| heterochromatin organization | 15 | 1.05 | 1.16 | 1.35 | 1.05 | 1.34 | 1.18 | 1.25 |
| heterochromatin maintenance | 1 | -1.2 | 1.03 | 1.07 | -1.11 | 1.52 | -1.28 | -1.06 |
| basement membrane assembly | 1 | -1.47 | 1.17 | 2.02 | 1.22 | 1.63 | -1.12 | 1.4 |
| divalent metal ion transport | 28 | 1.38 | -1.01 | 1.09 | -1.09 | 1.16 | 1.51 | 1.18 |
| growth factor receptor binding | 19 | 2.02 | -1.49 | -1.23 | -1.57 | 1.04 | 2.84 | 1.44 |
| myosin VI binding | 3 | -1.98 | 1.02 | 1.13 | 1.07 | 1.24 | -1.12 | -1.15 |
| myosin VI heavy chain binding | 2 | -1.4 | -1.01 | 1.0 | 1.02 | 1.17 | -1.17 | -1.2 |
| myosin VI head/neck binding | 2 | -1.4 | -1.01 | 1.0 | 1.02 | 1.17 | -1.17 | -1.2 |
| myosin VI light chain binding | 1 | -3.92 | 1.08 | 1.44 | 1.19 | 1.39 | -1.01 | -1.05 |
| regulation of protein exit from endoplasmic reticulum | 3 | -1.03 | 1.07 | -1.01 | 1.14 | -1.11 | -1.45 | 1.2 |
| positive regulation of protein exit from endoplasmic reticulum | 2 | -1.03 | 1.1 | -1.05 | 1.06 | -1.29 | -1.58 | 1.34 |
| sperm individualization complex | 8 | -1.42 | 1.11 | 1.46 | 1.09 | 1.37 | -1.03 | 1.02 |
| investment cone | 3 | -1.57 | 1.04 | 1.08 | 1.04 | 1.05 | -1.06 | -1.09 |
| heterochromatin organization involved in chromatin silencing | 6 | 1.24 | 1.15 | 1.45 | 1.05 | 1.33 | 1.19 | 1.28 |
| regulation of glycogen metabolic process | 1 | -2.86 | 1.25 | -1.2 | 1.45 | 1.26 | 1.29 | -1.32 |
| pre-miRNA binding | 1 | 1.6 | 1.11 | 4.34 | -1.01 | 3.45 | 5.33 | 3.25 |
| E-box binding | 1 | 1.39 | -1.11 | 1.11 | -1.25 | 1.17 | 1.75 | 1.6 |
| production of small RNA involved in gene silencing by RNA | 11 | 1.46 | 1.17 | 1.45 | 1.09 | 1.32 | 1.46 | 1.31 |
| regulation of production of small RNA involved in gene silencing by RNA | 2 | 2.75 | 1.11 | 1.02 | 1.03 | -1.06 | 1.3 | 1.12 |
| small RNA loading onto RISC | 6 | 1.25 | 1.16 | 1.42 | 1.05 | 1.43 | 1.57 | 1.49 |
| siRNA loading onto RISC involved in chromatin silencing by small RNA | 2 | 1.38 | 1.18 | -1.01 | 1.15 | 1.25 | 2.63 | 1.7 |
| organelle assembly | 50 | 1.16 | 1.06 | 1.16 | 1.03 | 1.21 | 1.22 | 1.1 |
| contractile ring | 5 | -1.53 | 1.2 | 1.44 | 1.14 | 1.64 | 1.07 | 1.08 |
| dephosphorylation of RNA polymerase II C-terminal domain | 1 | 1.1 | 1.03 | 1.12 | 1.08 | 1.22 | -1.0 | 1.51 |
| endoplasmic reticulum exit site | 1 | -2.09 | 1.34 | 1.35 | 1.01 | 1.51 | -1.24 | 2.45 |
| protein localization in endoplasmic reticulum | 18 | -1.3 | 1.35 | -1.06 | 1.05 | -1.19 | -1.62 | 1.27 |
| dendrite guidance | 17 | 1.16 | 1.07 | 1.26 | 1.05 | 1.37 | 1.01 | 1.27 |
| SET domain binding | 1 | 1.32 | 1.05 | 1.07 | 1.15 | 1.03 | -1.21 | -1.15 |
| left/right axis specification | 1 | -1.92 | 1.09 | 4.17 | -1.09 | 2.38 | -1.63 | 1.77 |
| demethylation | 7 | 1.18 | 1.25 | 1.72 | 1.13 | 1.59 | 1.68 | 1.3 |
| translation preinitiation complex | 1 | 2.17 | 1.18 | 1.52 | -1.1 | 1.33 | 2.16 | 1.32 |
| neuron death | 2 | -1.07 | 1.32 | 1.73 | -1.0 | -1.14 | 1.12 | 1.39 |
| response to magnetism | 1 | 1.37 | -1.11 | -1.04 | 1.15 | 1.03 | 2.71 | 1.13 |
| precatalytic spliceosome | 137 | 1.14 | 1.1 | 1.21 | 1.22 | 1.3 | 1.14 | -1.01 |
| catalytic step 2 spliceosome | 111 | 1.16 | 1.09 | 1.21 | 1.23 | 1.31 | 1.15 | -1.02 |
| RNA surveillance | 3 | 1.11 | 1.23 | 1.25 | 1.23 | 1.33 | 1.3 | 1.34 |
| nuclear RNA surveillance | 3 | 1.11 | 1.23 | 1.25 | 1.23 | 1.33 | 1.3 | 1.34 |
| histone mRNA catabolic process | 1 | 1.7 | -1.05 | -1.07 | -1.09 | -1.18 | 1.33 | 1.14 |
| DNA conformation change | 78 | 1.1 | 1.1 | 1.24 | 1.13 | 1.27 | 1.09 | 1.11 |
| regulation of cell cycle arrest | 84 | 1.23 | 1.14 | 1.35 | 1.19 | 1.41 | 1.29 | 1.15 |
| ribonucleoprotein complex localization | 1 | 2.15 | -1.01 | 1.44 | 1.25 | 1.44 | 1.28 | 1.21 |
| spindle assembly checkpoint | 10 | 1.07 | 1.09 | 1.16 | 1.15 | 1.32 | 1.1 | 1.03 |
| mitotic cell cycle spindle checkpoint | 10 | 1.07 | 1.09 | 1.16 | 1.15 | 1.32 | 1.1 | 1.03 |
| subsynaptic reticulum | 1 | 4.69 | -1.04 | -1.32 | -1.4 | -1.38 | 1.37 | -1.05 |
| cellular response to abiotic stimulus | 19 | -1.22 | -1.34 | -1.35 | -1.33 | -1.19 | 1.12 | -1.02 |
| cellular response to biotic stimulus | 4 | -1.08 | 1.29 | 1.46 | 1.13 | 1.58 | 1.05 | 1.4 |
| cellular response to molecule of bacterial origin | 2 | 1.04 | 1.17 | 1.37 | 1.17 | 1.75 | 1.28 | 1.1 |
| cellular response to lipopolysaccharide | 2 | 1.04 | 1.17 | 1.37 | 1.17 | 1.75 | 1.28 | 1.1 |
| cellular response to acid | 5 | 1.44 | -1.04 | 1.24 | 1.2 | 1.26 | 1.13 | -1.15 |
| cellular response to inorganic substance | 8 | 1.06 | 1.09 | 1.5 | 1.11 | 1.42 | 1.08 | 1.06 |
| cellular response to arsenic-containing substance | 6 | 1.03 | 1.11 | 1.5 | 1.16 | 1.48 | 1.0 | 1.06 |
| cellular response to carbon dioxide | 2 | 1.12 | 1.05 | 1.51 | -1.02 | 1.24 | 1.35 | 1.06 |
| cellular response to metal ion | 6 | 1.03 | 1.11 | 1.5 | 1.16 | 1.48 | 1.0 | 1.06 |
| cytoplasmic U snRNP body | 6 | 1.33 | -1.0 | 1.21 | 1.26 | 1.29 | 1.29 | -1.06 |
| translocon complex | 8 | -1.51 | 1.3 | -1.14 | -1.0 | -1.28 | -1.7 | 1.22 |
| L-methionine biosynthetic process | 2 | 1.35 | 1.12 | -1.14 | 1.23 | 1.0 | 1.41 | -1.09 |
| L-methionine salvage | 2 | 1.35 | 1.12 | -1.14 | 1.23 | 1.0 | 1.41 | -1.09 |
| cellular response to cadmium ion | 6 | 1.03 | 1.11 | 1.5 | 1.16 | 1.48 | 1.0 | 1.06 |
| cellular response to organic substance | 81 | 1.21 | 1.09 | 1.31 | 1.07 | 1.23 | 1.27 | 1.19 |
| cellular response to dsRNA | 11 | 1.46 | 1.17 | 1.45 | 1.09 | 1.32 | 1.46 | 1.31 |
| cellular response to ethanol | 1 | 1.36 | 1.32 | 2.03 | 1.27 | 1.67 | 1.42 | 1.61 |
| cellular response to growth factor stimulus | 12 | 1.59 | -1.08 | 1.97 | -1.01 | 1.17 | 1.52 | 1.1 |
| cellular response to peptide hormone stimulus | 20 | -1.09 | 1.13 | 1.2 | 1.13 | 1.18 | 1.07 | 1.2 |
| cellular response to steroid hormone stimulus | 29 | 1.17 | 1.11 | 1.15 | 1.05 | 1.22 | 1.27 | 1.21 |
| cellular response to organic nitrogen | 4 | 1.59 | -1.01 | 1.38 | 1.29 | 1.34 | 1.2 | -1.18 |
| cellular response to amine stimulus | 4 | 1.59 | -1.01 | 1.38 | 1.29 | 1.34 | 1.2 | -1.18 |
| succinate transmembrane transport | 1 | -1.02 | -2.78 | 1.86 | -1.91 | 2.75 | 13.01 | -1.81 |
| malate transmembrane transport | 2 | 1.03 | -1.5 | 1.48 | -1.27 | 1.66 | 3.63 | -1.34 |
| ribonucleoprotein complex export from nucleus | 1 | 2.15 | -1.01 | 1.44 | 1.25 | 1.44 | 1.28 | 1.21 |
| rRNA-containing ribonucleoprotein complex export from nucleus | 1 | 2.15 | -1.01 | 1.44 | 1.25 | 1.44 | 1.28 | 1.21 |
| cellular response to oxygen radical | 2 | 1.69 | 1.17 | 1.01 | 1.09 | 1.11 | 1.41 | -1.08 |
| cellular response to superoxide | 2 | 1.69 | 1.17 | 1.01 | 1.09 | 1.11 | 1.41 | -1.08 |
| cellular response to oxygen levels | 25 | 1.18 | 1.19 | 1.57 | 1.15 | 1.38 | 1.41 | 1.17 |
| cellular response to anoxia | 2 | -1.34 | -1.15 | -1.19 | 1.14 | -1.2 | -1.27 | -1.19 |
| cellular response to hypoxia | 23 | 1.23 | 1.22 | 1.66 | 1.15 | 1.44 | 1.48 | 1.21 |
| protein localization to chromosome, centromeric region | 1 | 2.47 | 1.54 | 1.18 | 1.04 | 1.48 | 1.78 | 1.43 |
| cellular response to water | 1 | -1.22 | -1.01 | 1.11 | -1.0 | -1.07 | -1.09 | 1.17 |
| cellular response to xenobiotic stimulus | 10 | 1.1 | -1.02 | -1.79 | -1.06 | -1.28 | -1.46 | 1.06 |
| cellular response to osmotic stress | 2 | 1.01 | 1.17 | 1.53 | -1.02 | 1.61 | 1.26 | 1.23 |
| cellular response to salt stress | 1 | 1.22 | 1.33 | 2.18 | 1.07 | 1.72 | 2.05 | 1.61 |
| cellular hyperosmotic response | 1 | 1.22 | 1.33 | 2.18 | 1.07 | 1.72 | 2.05 | 1.61 |
| cellular hyperosmotic salinity response | 1 | 1.22 | 1.33 | 2.18 | 1.07 | 1.72 | 2.05 | 1.61 |
| cellular response to radiation | 16 | -1.25 | -1.45 | -1.52 | -1.39 | -1.3 | 1.12 | -1.06 |
| cellular response to ionizing radiation | 1 | -1.03 | 1.12 | 1.58 | 1.2 | 2.16 | 1.52 | 1.14 |
| cellular response to X-ray | 1 | -1.03 | 1.12 | 1.58 | 1.2 | 2.16 | 1.52 | 1.14 |
| cellular response to light stimulus | 15 | -1.27 | -1.49 | -1.61 | -1.44 | -1.39 | 1.09 | -1.07 |
| cellular response to light intensity | 1 | 3.41 | -1.85 | -1.6 | -1.39 | -1.6 | 6.88 | -1.28 |
| cellular response to high light intensity | 1 | 3.41 | -1.85 | -1.6 | -1.39 | -1.6 | 6.88 | -1.28 |
| cellular response to endogenous stimulus | 68 | 1.18 | 1.07 | 1.29 | 1.07 | 1.22 | 1.24 | 1.16 |
| cellular response to external stimulus | 17 | -1.03 | -1.05 | -1.04 | -1.13 | 1.01 | 1.48 | 1.15 |
| cellular response to sterol depletion | 1 | 1.18 | 1.17 | 1.43 | 1.18 | 1.36 | -1.03 | 1.42 |
| genetic imprinting | 1 | 1.01 | 1.1 | 1.09 | 1.14 | -1.01 | 1.17 | 1.11 |
| semaphorin-plexin signaling pathway | 2 | 1.11 | -1.03 | 1.73 | -1.06 | 1.28 | 1.08 | 1.11 |
| inositol phosphate catabolic process | 3 | 2.42 | 1.04 | 1.61 | -1.01 | 1.39 | 1.64 | 1.29 |
| cell wall organization or biogenesis | 9 | -1.18 | -1.16 | -1.42 | -1.31 | -1.28 | -2.35 | -1.08 |
| histone H3-K27 demethylation | 1 | 1.79 | 1.29 | 2.63 | 1.3 | 1.91 | 2.12 | 1.41 |
| response to transforming growth factor beta stimulus | 2 | 1.38 | 1.12 | 1.33 | 1.08 | 1.33 | 1.38 | 1.15 |
| cellular response to transforming growth factor beta stimulus | 2 | 1.38 | 1.12 | 1.33 | 1.08 | 1.33 | 1.38 | 1.15 |
| acyl-CoA biosynthetic process | 5 | 2.3 | 1.04 | 1.28 | 1.15 | 1.53 | 1.22 | 1.09 |
| lysophospholipid acyltransferase activity | 4 | 1.01 | 1.34 | 1.66 | -1.03 | 1.72 | 1.33 | 1.31 |
| optomotor response | 2 | 1.16 | 1.09 | 1.72 | 1.19 | 1.37 | 1.2 | 2.11 |
| Slit-Robo signaling complex | 1 | 2.19 | 1.14 | 2.0 | 1.19 | 2.09 | 1.71 | 1.7 |
| sensory dendrite | 7 | -1.01 | 1.01 | 1.05 | 1.03 | -1.01 | 1.05 | -1.02 |
| organism emergence from protective structure | 18 | -1.05 | -1.01 | 1.01 | 1.0 | -1.05 | -1.13 | 1.1 |
| striated muscle myosin thick filament assembly | 4 | 2.6 | -1.28 | 1.1 | -1.18 | 1.07 | 1.9 | -1.04 |
| protein localization in extracellular region | 1 | 1.79 | 1.14 | -1.18 | 1.07 | 1.25 | -5.71 | 1.37 |
| anatomical structure maturation | 3 | 3.11 | -1.4 | 1.27 | -1.39 | 1.24 | 4.46 | 1.69 |
| ectodermal placode development | 4 | -1.14 | 1.1 | 1.23 | 1.12 | 1.47 | -1.2 | -1.08 |
| ectodermal placode morphogenesis | 4 | -1.14 | 1.1 | 1.23 | 1.12 | 1.47 | -1.2 | -1.08 |
| organic substance transport | 97 | -1.4 | -1.24 | -1.57 | -1.12 | -1.07 | -1.03 | -1.27 |
| nitrogen compound transport | 51 | -1.34 | -1.08 | -1.59 | -1.16 | 1.0 | -1.05 | -1.09 |
| basement membrane organization | 5 | -1.2 | 1.03 | 1.32 | -1.01 | 1.32 | 1.16 | 1.46 |
| detoxification of arsenic-containing substance | 1 | 1.35 | 1.29 | 1.36 | 1.08 | 1.19 | 1.34 | 1.11 |
| transcriptional activation by promoter-enhancer looping | 1 | -1.14 | 1.04 | 1.08 | -1.05 | -1.05 | -1.08 | 1.07 |
| response to BMP stimulus | 1 | -1.42 | 1.06 | 1.32 | 1.03 | 1.22 | -1.52 | 1.31 |
| response to fibroblast growth factor stimulus | 10 | 1.63 | -1.12 | 2.13 | -1.03 | 1.14 | 1.56 | 1.09 |
| regulation of cell cycle cytokinesis | 1 | -1.32 | 1.18 | 1.39 | 1.03 | 1.12 | 1.4 | 1.35 |
| G1/S transition checkpoint | 2 | 1.09 | 1.49 | 1.71 | 1.02 | 1.44 | 1.9 | 1.73 |
| mitotic cell cycle G2/M transition checkpoint | 62 | 1.22 | 1.15 | 1.36 | 1.19 | 1.38 | 1.27 | 1.14 |
| cellular potassium ion transport | 10 | 1.06 | 1.08 | 1.11 | -1.23 | -1.25 | 1.17 | -1.26 |
| potassium ion transmembrane transport | 10 | 1.06 | 1.08 | 1.11 | -1.23 | -1.25 | 1.17 | -1.26 |
| protein transmembrane transport | 5 | -1.62 | 1.28 | -1.17 | -1.01 | -1.28 | -1.9 | 1.13 |
| protein complex subunit organization | 94 | -1.0 | 1.1 | 1.26 | 1.11 | 1.25 | 1.13 | -1.0 |
| protein-DNA complex subunit organization | 47 | 1.05 | 1.08 | 1.28 | 1.18 | 1.29 | -1.03 | 1.03 |
| ribonucleoprotein complex subunit organization | 34 | 1.25 | 1.14 | 1.3 | 1.19 | 1.3 | 1.25 | 1.1 |
| cellular component organization or biogenesis | 1726 | 1.17 | 1.08 | 1.21 | 1.06 | 1.23 | 1.21 | 1.12 |
| cellular component organization or biogenesis at cellular level | 1496 | 1.17 | 1.07 | 1.22 | 1.07 | 1.24 | 1.21 | 1.12 |
| cellular component organization at cellular level | 1453 | 1.17 | 1.07 | 1.22 | 1.06 | 1.24 | 1.21 | 1.13 |
| cellular component biogenesis at cellular level | 87 | 1.26 | 1.15 | 1.28 | 1.23 | 1.29 | 1.18 | 1.07 |
| cellular component assembly at cellular level | 446 | 1.19 | 1.06 | 1.24 | 1.05 | 1.23 | 1.23 | 1.12 |
| cellular component disassembly at cellular level | 27 | -1.1 | 1.18 | 1.3 | 1.14 | 1.29 | 1.09 | 1.04 |
| adrenergic receptor signaling pathway | 2 | -1.09 | 1.2 | -1.08 | -1.0 | -1.18 | -1.21 | -1.07 |
| DNA biosynthetic process | 25 | 1.09 | 1.18 | 1.33 | 1.16 | 1.4 | 1.14 | 1.1 |
| regulation of protein serine/threonine kinase activity | 30 | 1.29 | 1.16 | 1.42 | 1.17 | 1.4 | 1.39 | 1.17 |
| negative regulation of protein serine/threonine kinase activity | 7 | 1.54 | 1.14 | 1.5 | 1.16 | 1.5 | 1.83 | 1.37 |
| positive regulation of protein serine/threonine kinase activity | 9 | 1.14 | 1.19 | 1.51 | 1.11 | 1.38 | 1.84 | 1.15 |
| octopamine signaling pathway | 1 | 1.3 | 1.0 | -1.05 | -1.09 | 1.04 | -1.06 | -1.0 |
| nitrogen cycle metabolic process | 1 | -31.89 | 1.77 | -20.72 | 1.41 | 1.0 | -12.66 | -3.5 |
| cell periphery | 658 | 1.15 | 1.02 | 1.13 | -1.02 | 1.12 | 1.22 | 1.15 |
| conversion of methionyl-tRNA to N-formyl-methionyl-tRNA | 1 | 1.38 | 1.09 | 1.78 | 1.2 | 1.53 | 2.7 | -1.25 |
| cellular component maintenance at cellular level | 12 | -1.17 | 1.2 | 1.56 | 1.1 | 1.45 | 1.12 | 1.1 |
| renal system development | 69 | 1.2 | -1.03 | 1.12 | -1.01 | 1.21 | 1.35 | 1.08 |
| Malpighian tubule development | 62 | 1.15 | -1.03 | 1.11 | -1.01 | 1.22 | 1.34 | 1.08 |
| nephron development | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| nephron epithelium development | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| nephron morphogenesis | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| kidney epithelium development | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| nephron tubule morphogenesis | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| nephron tubule formation | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| nephron tubule development | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| nephron epithelium morphogenesis | 3 | -1.1 | -1.05 | -1.04 | -1.15 | -1.04 | 1.01 | 1.54 |
| stem cell proliferation | 5 | 1.52 | 1.24 | 1.48 | 1.18 | 1.44 | 1.13 | 1.17 |
| regulation of stem cell proliferation | 2 | -1.24 | 1.36 | -1.02 | -1.09 | 1.25 | 1.32 | 3.25 |
| epithelial cell fate commitment | 3 | 1.0 | 1.08 | 1.89 | 1.0 | 1.21 | -1.44 | -1.04 |
| epithelial tube formation | 6 | 1.03 | -1.09 | 1.01 | -1.18 | -1.08 | 1.45 | 1.48 |
| vesicle uncoating | 2 | -1.44 | 1.22 | 1.71 | 1.06 | 1.91 | 1.53 | 1.42 |
| volume-sensitive chloride channel activity | 1 | 3.44 | -1.08 | -1.56 | -1.91 | 1.81 | 5.18 | 1.78 |
| chaperone-mediated protein transport | 1 | 1.09 | 1.09 | 1.08 | -1.0 | -1.04 | -1.07 | -1.01 |
| monocarboxylic acid catabolic process | 15 | -1.0 | 1.02 | -1.46 | 1.02 | 1.13 | -1.74 | -1.3 |
| monocarboxylic acid biosynthetic process | 2 | 1.14 | 1.17 | -1.17 | -1.01 | 1.02 | 1.07 | -1.0 |
| signal transduction by p53 class mediator | 2 | 1.7 | 1.37 | 1.03 | 1.24 | 1.62 | 1.04 | 1.2 |
| signal transduction by p53 class mediator resulting in induction of apoptosis | 2 | 1.7 | 1.37 | 1.03 | 1.24 | 1.62 | 1.04 | 1.2 |
| cellular lactam metabolic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| cellular lactam biosynthetic process | 1 | -1.31 | -1.76 | 1.72 | 1.22 | 1.97 | 3.11 | 1.28 |
| modified amino acid binding | 5 | -1.4 | -1.12 | -1.47 | 1.21 | 1.21 | -1.15 | -1.62 |
| response to anesthetic | 7 | 1.2 | 1.01 | 1.0 | 1.08 | 1.05 | 1.41 | -1.12 |
| sulfur compound transport | 2 | -2.47 | -1.07 | -3.17 | 1.42 | 1.77 | -1.97 | -2.08 |
| modified amino acid transmembrane transporter activity | 1 | -7.18 | -1.45 | -9.47 | 1.62 | 3.49 | -2.41 | -5.84 |
| tricarboxylic acid metabolic process | 2 | -1.17 | -1.01 | -1.18 | 1.04 | -1.26 | -1.29 | -1.1 |
| cardiovascular system development | 70 | 1.28 | -1.02 | 1.19 | -1.07 | 1.17 | 1.3 | 1.17 |
| primary cilium | 2 | -1.07 | -1.03 | -1.02 | 1.13 | -1.1 | -1.09 | -1.15 |
| medium-term memory | 5 | -2.06 | 1.08 | 1.11 | 1.07 | -1.16 | -1.78 | -1.05 |
| organelle transport along microtubule | 7 | -1.28 | 1.21 | 1.46 | 1.16 | 1.42 | 1.38 | 1.37 |
| microtubule anchoring at microtubule organizing center | 1 | -1.51 | -1.02 | 1.38 | 1.04 | 1.42 | 1.11 | -1.1 |
| ammonium transmembrane transport | 1 | -1.13 | -1.02 | -1.13 | -1.06 | -1.15 | -1.01 | -1.16 |
| photoreceptor cell axon guidance | 3 | -1.41 | 1.34 | 1.88 | 1.12 | 1.32 | 1.49 | 1.42 |
| divalent inorganic cation transmembrane transporter activity | 19 | -1.37 | 1.2 | 1.22 | 1.05 | 1.11 | -1.13 | -1.04 |
| divalent inorganic cation transport | 29 | 1.35 | 1.0 | 1.1 | -1.08 | 1.16 | 1.48 | 1.17 |
| purine-containing compound metabolic process | 132 | 1.15 | 1.03 | 1.11 | 1.02 | 1.08 | 1.22 | -1.04 |
| purine-containing compound biosynthetic process | 74 | 1.06 | 1.04 | 1.09 | 1.01 | 1.04 | 1.27 | -1.07 |
| purine-containing compound catabolic process | 40 | 1.18 | 1.04 | 1.15 | 1.09 | 1.13 | 1.08 | 1.01 |
| pyridine-containing compound metabolic process | 11 | 1.88 | 1.17 | -1.48 | -1.05 | 1.08 | 1.25 | 1.36 |
| pyridine-containing compound biosynthetic process | 3 | 1.9 | 1.5 | -1.09 | 1.01 | 1.09 | 1.23 | 1.86 |
| pyrimidine-containing compound metabolic process | 22 | 1.04 | 1.05 | 1.04 | 1.17 | 1.19 | -1.08 | -1.06 |
| pyrimidine-containing compound biosynthetic process | 14 | 1.27 | 1.04 | 1.16 | 1.11 | 1.17 | 1.08 | -1.03 |
| pyrimidine-containing compound catabolic process | 6 | 1.02 | -1.16 | -1.31 | -1.06 | -1.01 | -1.14 | 1.05 |
| purine-containing compound transmembrane transport | 5 | -1.03 | 1.21 | 1.18 | 1.23 | 1.16 | 1.19 | -1.13 |
| pyrimidine-containing compound transmembrane transport | 4 | -1.46 | 1.25 | 1.0 | 1.26 | 1.04 | -1.57 | 1.43 |
| protein phosphatase activator activity | 3 | 1.39 | 1.03 | 1.11 | 1.1 | 1.16 | 1.44 | 1.03 |
| ADP-D-ribose binding | 1 | 1.71 | 1.0 | 1.2 | 1.14 | 1.43 | 1.37 | 1.01 |
| poly-ADP-D-ribose binding | 1 | 1.71 | 1.0 | 1.2 | 1.14 | 1.43 | 1.37 | 1.01 |
| clathrin-mediated endocytosis | 1 | 4.7 | -1.51 | -1.0 | -1.26 | 1.19 | 8.29 | 2.05 |
| DNA topoisomerase (ATP-hydrolyzing) regulator activity | 1 | 1.69 | 1.07 | 1.4 | 1.09 | 1.95 | 1.43 | 1.53 |
| DNA topoisomerase (ATP-hydrolyzing) activator activity | 1 | 1.69 | 1.07 | 1.4 | 1.09 | 1.95 | 1.43 | 1.53 |
| box H/ACA RNP complex | 1 | 1.3 | 1.21 | 1.13 | 1.29 | 1.21 | 1.06 | 1.05 |
| establishment of protein localization to organelle | 60 | 1.01 | 1.17 | 1.14 | 1.11 | 1.09 | 1.02 | 1.1 |
| maintenance of protein localization to organelle | 4 | -1.56 | 1.22 | -1.02 | 1.14 | -1.0 | -1.78 | 1.25 |
| establishment of protein localization in endoplasmic reticulum | 15 | -1.21 | 1.37 | -1.05 | 1.04 | -1.19 | -1.54 | 1.26 |
| establishment of protein localization in Golgi | 10 | -1.16 | 1.12 | 1.17 | 1.04 | 1.21 | 1.41 | 1.65 |
| establishment of protein localization in mitochondrion | 28 | 1.07 | 1.09 | 1.21 | 1.13 | 1.12 | 1.18 | -1.1 |
| protein localization to peroxisome | 3 | 1.5 | 1.17 | -1.6 | 1.44 | 1.46 | -2.19 | -1.64 |
| establishment of protein localization to peroxisome | 3 | 1.5 | 1.17 | -1.6 | 1.44 | 1.46 | -2.19 | -1.64 |
| protein localization to vacuole | 4 | 1.47 | 1.26 | 2.02 | 1.17 | 1.52 | 1.6 | 1.47 |
| establishment of protein localization to vacuole | 4 | 1.47 | 1.26 | 2.02 | 1.17 | 1.52 | 1.6 | 1.47 |
| tubulin complex biogenesis | 4 | 1.19 | 1.1 | 1.33 | 1.15 | 1.38 | -1.02 | 1.05 |
| mitotic spindle | 14 | 1.25 | 1.22 | 1.39 | 1.19 | 1.29 | 1.28 | 1.12 |
| meiotic spindle | 1 | -1.24 | -1.06 | -1.4 | -1.21 | -1.33 | -1.23 | -1.12 |
| S-nitrosoglutathione reductase activity | 1 | 1.29 | 1.1 | 1.03 | 1.22 | 1.29 | 1.66 | -1.14 |
| regulation of reactive oxygen species metabolic process | 8 | 1.29 | 1.23 | 1.39 | 1.25 | 1.52 | 1.58 | 1.24 |
| 3R-hydroxyacyl-CoA dehydratase activity | 1 | -1.23 | 1.06 | -2.67 | 1.58 | 1.11 | -5.4 | -1.98 |
| regulation of primary metabolic process | 957 | 1.15 | 1.09 | 1.2 | 1.07 | 1.19 | 1.19 | 1.13 |
| regulation of response to stress | 72 | 1.36 | -1.02 | 1.09 | 1.0 | 1.13 | 1.39 | 1.15 |
| regulation of cellular response to stress | 35 | -1.02 | 1.21 | 1.29 | 1.13 | 1.3 | 1.36 | 1.16 |
| regulation of nitric oxide metabolic process | 1 | 1.29 | 1.1 | 1.03 | 1.22 | 1.29 | 1.66 | -1.14 |
| histone H3-K4 trimethylation | 1 | 1.69 | 1.2 | 1.93 | 1.03 | 2.02 | 2.04 | 1.95 |
| extracellular matrix assembly | 12 | 1.14 | 1.01 | -1.05 | -1.02 | -1.08 | 1.01 | -1.0 |
| regulation of mitotic anaphase | 1 | 1.43 | 1.15 | 2.06 | 1.04 | 1.96 | 1.44 | 1.33 |
| regulation of chromatin silencing at centromere | 1 | 1.21 | 1.08 | 1.29 | -1.08 | 1.41 | 1.2 | 1.17 |
| positive regulation of chromatin silencing at centromere | 1 | 1.21 | 1.08 | 1.29 | -1.08 | 1.41 | 1.2 | 1.17 |
| regulation of trehalose metabolic process | 1 | 1.03 | -1.29 | -2.06 | -2.17 | -1.97 | -2.33 | 4.44 |
| positive regulation of microtubule nucleation | 1 | 2.49 | 1.3 | 2.87 | -1.0 | 2.26 | 3.13 | 2.77 |
| regulation of anatomical structure size | 118 | 1.38 | 1.02 | 1.24 | -1.04 | 1.24 | 1.48 | 1.3 |
| positive regulation of cell cycle process | 13 | 1.26 | 1.06 | 1.08 | 1.01 | 1.3 | 1.62 | 1.07 |
| regulation of ribosome biogenesis | 2 | 1.63 | 1.23 | 1.65 | 1.22 | 1.61 | 2.39 | 1.43 |
| positive regulation of ribosome biogenesis | 2 | 1.63 | 1.23 | 1.65 | 1.22 | 1.61 | 2.39 | 1.43 |
| positive regulation of sodium ion transport via voltage-gated sodium channel activity | 5 | 1.02 | 1.03 | 1.03 | 1.0 | -1.03 | 1.03 | -1.06 |
| translation regulator activity, nucleic acid binding | 7 | 1.85 | 1.07 | 2.03 | 1.03 | 1.61 | 1.55 | 1.78 |
| regulation of peptide transport | 1 | 1.12 | 1.15 | 1.24 | 1.0 | 1.0 | 1.1 | -1.06 |
| regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 23 | 1.18 | 1.12 | 1.35 | 1.09 | 1.3 | 1.19 | 1.21 |
| regulation of decapentaplegic signaling pathway | 6 | 1.3 | 1.01 | 1.61 | -1.03 | 1.09 | 1.53 | 1.36 |
| positive regulation of decapentaplegic signaling pathway | 1 | 1.81 | 1.21 | 1.48 | 1.18 | 1.37 | 1.47 | 1.47 |
| negative regulation of decapentaplegic signaling pathway | 3 | 1.28 | -1.1 | 1.11 | -1.15 | -1.09 | 1.84 | 1.01 |
| positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 5 | 1.47 | 1.09 | 1.08 | -1.01 | 1.15 | 1.29 | 1.21 |
| negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 13 | 1.13 | 1.1 | 1.3 | 1.09 | 1.27 | 1.11 | 1.16 |
| regulation of cell-substrate junction assembly | 1 | 2.96 | 1.13 | 1.75 | -1.2 | 1.98 | 1.94 | 1.75 |
| COPII-coated vesicle budding | 1 | -1.49 | 1.52 | 1.25 | 1.12 | 1.19 | 1.46 | 1.78 |
| tissue migration | 36 | 1.66 | -1.04 | 1.31 | -1.09 | 1.2 | 1.63 | 1.1 |
| epithelium migration | 28 | 1.66 | -1.03 | 1.29 | -1.07 | 1.21 | 1.38 | 1.11 |
| regulation of mitochondrial fission | 2 | -1.0 | 1.14 | -1.27 | 1.12 | -1.13 | -1.52 | -1.35 |
| positive regulation of mitochondrial fission | 2 | -1.0 | 1.14 | -1.27 | 1.12 | -1.13 | -1.52 | -1.35 |
| endoplasmic reticulum membrane organization | 1 | -1.04 | 1.0 | 1.09 | 1.3 | 1.23 | -1.23 | -1.03 |
| establishment of epithelial cell polarity | 11 | 1.26 | 1.04 | 1.35 | -1.0 | 1.51 | 1.34 | 1.27 |
| establishment of epithelial cell planar polarity | 4 | -1.26 | 1.08 | 1.52 | 1.16 | 1.36 | 1.11 | -1.02 |
| organelle membrane fusion | 4 | 1.13 | 1.23 | 1.24 | 1.17 | 1.16 | 1.0 | 1.22 |
| regulation of establishment of planar polarity | 4 | 1.06 | -1.1 | 1.26 | -1.16 | 1.25 | 1.67 | 1.42 |
| microtubule cytoskeleton organization involved in establishment of planar polarity | 3 | 1.57 | 1.03 | 1.21 | -1.01 | 1.13 | 2.03 | 1.55 |
| gene looping | 1 | -1.14 | 1.04 | 1.08 | -1.05 | -1.05 | -1.08 | 1.07 |
| regulation of triglyceride metabolic process | 2 | -1.95 | -1.1 | -1.54 | 1.16 | 1.88 | -3.69 | -2.32 |
| regulation of spindle organization | 10 | 1.06 | 1.17 | 1.27 | 1.15 | 1.35 | 1.28 | 1.1 |
| regulation of spindle checkpoint | 1 | 1.18 | -1.09 | 1.06 | 1.12 | 1.44 | -1.15 | -1.22 |
| regulation of histone H4 acetylation | 1 | 1.8 | 1.23 | 1.54 | 1.26 | 1.37 | 1.4 | 1.76 |
| positive regulation of histone H4 acetylation | 1 | 1.8 | 1.23 | 1.54 | 1.26 | 1.37 | 1.4 | 1.76 |
| cell-cell adhesion involved in establishment of planar polarity | 1 | 1.3 | 1.94 | 1.14 | 1.03 | 1.07 | 1.88 | 1.31 |
| cell elongation involved in imaginal disc-derived wing morphogenesis | 5 | 1.41 | 1.11 | 1.92 | 1.05 | 1.98 | 1.8 | 1.61 |
| regulation of muscle system process | 5 | 2.39 | -1.02 | -2.05 | -2.54 | -2.17 | 1.16 | 1.24 |
| positive regulation of canonical Wnt receptor signaling pathway | 4 | 1.23 | 1.08 | 1.27 | 1.09 | -1.0 | 1.0 | 1.22 |
| regulation of calcium ion import | 1 | -1.32 | 1.16 | 1.21 | -1.14 | -1.21 | 1.47 | 1.08 |
| negative regulation of calcium ion import | 1 | -1.32 | 1.16 | 1.21 | -1.14 | -1.21 | 1.47 | 1.08 |
| nucleic acid metabolic process | 788 | 1.2 | 1.12 | 1.23 | 1.17 | 1.28 | 1.17 | 1.07 |
| nucleic acid phosphodiester bond hydrolysis | 23 | 1.3 | 1.21 | 1.39 | 1.25 | 1.38 | 1.28 | 1.18 |
| spindle assembly involved in meiosis | 13 | -1.1 | 1.07 | 1.06 | 1.07 | 1.24 | -1.2 | -1.03 |
| spindle assembly involved in mitosis | 3 | -1.0 | 1.13 | 1.24 | 1.35 | 1.53 | -1.14 | -1.07 |
| regulation of protein deacetylation | 1 | 1.03 | 1.14 | 1.11 | 1.37 | 1.92 | -1.3 | 1.15 |
| positive regulation of protein deacetylation | 1 | 1.03 | 1.14 | 1.11 | 1.37 | 1.92 | -1.3 | 1.15 |
| regulation of protein targeting to membrane | 1 | 1.09 | 1.09 | 1.08 | -1.0 | -1.04 | -1.07 | -1.01 |
| positive regulation of protein targeting to membrane | 1 | 1.09 | 1.09 | 1.08 | -1.0 | -1.04 | -1.07 | -1.01 |
| positive regulation of intracellular protein transport | 23 | 1.1 | 1.07 | 1.2 | 1.09 | 1.25 | 1.08 | 1.14 |
| negative regulation of intracellular protein transport | 6 | 2.03 | 1.13 | 1.29 | 1.08 | 1.28 | 1.86 | 1.59 |
| regulation of olfactory learning | 2 | 1.05 | 1.07 | 1.07 | 1.08 | -1.09 | 1.03 | -1.03 |
| regulation of DNA-dependent DNA replication | 6 | -1.5 | 1.09 | 1.13 | 1.11 | 1.33 | -1.52 | 1.28 |
| regulation of heart induction | 3 | 1.09 | 1.1 | 1.13 | 1.08 | 1.18 | 1.14 | 2.09 |
| phagosome maturation | 1 | 1.76 | 1.27 | 1.35 | 1.51 | 1.34 | 1.49 | 1.23 |
| organophosphate biosynthetic process | 3 | -1.14 | 1.2 | -1.03 | -1.06 | 1.01 | -1.08 | 1.11 |
| ceramide binding | 1 | -2.39 | 1.47 | 1.51 | 1.07 | -1.25 | -2.16 | 1.24 |
| adipokinetic hormone receptor activity | 2 | 1.14 | 1.03 | 1.06 | 1.02 | -1.01 | -1.0 | -1.03 |
| adipokinetic hormone binding | 2 | 1.14 | 1.03 | 1.06 | 1.02 | -1.01 | -1.0 | -1.03 |
| adipokinetic hormone receptor binding | 1 | 2.14 | -13.11 | -13.63 | -12.76 | -16.87 | -14.38 | 5.61 |
| energy homeostasis | 1 | 1.86 | -1.04 | 1.59 | -1.1 | 2.08 | 2.71 | 1.26 |
| microtubule-based flagellar cytoplasm | 1 | 1.03 | 1.24 | 1.01 | 1.09 | 1.02 | -1.11 | 1.01 |
| mitochondrial respiratory chain complex I biogenesis | 1 | 1.08 | 1.05 | 1.31 | 1.03 | 1.07 | 1.99 | -1.52 |
| mitochondrial respiratory chain complex III biogenesis | 2 | 1.1 | 1.08 | 1.56 | 1.1 | 1.12 | 1.34 | -1.21 |
| regulation of membrane lipid distribution | 2 | 2.57 | -1.14 | 1.03 | -1.07 | 2.13 | 1.16 | 1.16 |
| perinuclear endoplasmic reticulum | 2 | -1.1 | 1.15 | 1.07 | 1.1 | 1.01 | 1.07 | 1.17 |
| synaptic membrane | 35 | -1.01 | 1.01 | 1.06 | -1.11 | -1.05 | 1.13 | 1.13 |
| dendritic spine organization | 1 | -2.52 | 1.99 | 1.32 | 1.32 | 1.09 | -1.34 | 1.43 |
| dendritic spine maintenance | 1 | -2.52 | 1.99 | 1.32 | 1.32 | 1.09 | -1.34 | 1.43 |
| anterior head development | 9 | 1.09 | -1.1 | 1.06 | 1.06 | -1.02 | -1.13 | -1.01 |
| pre-mRNA intronic binding | 1 | 1.28 | 1.07 | 1.73 | 1.12 | 1.48 | 1.94 | 1.41 |
| activation of cysteine-type endopeptidase activity | 13 | 1.0 | 1.15 | 1.25 | 1.21 | 1.45 | 1.21 | 1.07 |
| renal filtration | 2 | -1.2 | -1.04 | -1.19 | 1.02 | -1.08 | -1.21 | -1.15 |
| nephrocyte filtration | 2 | -1.2 | -1.04 | -1.19 | 1.02 | -1.08 | -1.21 | -1.15 |
| cell-type specific apoptotic process | 15 | 1.32 | 1.04 | 1.28 | -1.05 | 1.01 | 1.08 | 1.27 |
| regulation of cellular response to insulin stimulus | 10 | 1.25 | 1.09 | 1.37 | -1.04 | 1.17 | 1.16 | 1.59 |
| negative regulation of cellular response to insulin stimulus | 7 | 1.27 | 1.12 | 1.56 | -1.01 | 1.31 | 1.27 | 1.5 |
| positive regulation of cellular response to insulin stimulus | 2 | 1.0 | -1.0 | -1.17 | -1.4 | -1.23 | -1.17 | 2.26 |
| regulation of defense response to fungus | 7 | 2.0 | -1.32 | -1.17 | 1.03 | 1.03 | 1.92 | 1.18 |
| regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 4 | 1.18 | 1.11 | 1.21 | 1.11 | 1.04 | 1.21 | 1.05 |
| regulation of protein localization to nucleus | 30 | 1.19 | 1.1 | 1.26 | 1.1 | 1.28 | 1.31 | 1.24 |
| regulation of phospholipase C activity | 6 | 1.13 | 1.12 | 1.17 | 1.04 | 1.09 | 1.14 | 1.05 |
| regulation of multicellular organismal development | 278 | 1.14 | 1.08 | 1.25 | 1.05 | 1.25 | 1.23 | 1.17 |
| regulation of organ morphogenesis | 55 | 1.11 | 1.02 | 1.24 | 1.04 | 1.26 | 1.08 | 1.17 |
| regulation of planar cell polarity pathway involved in axis elongation | 2 | -1.01 | 1.74 | 1.54 | 1.1 | 1.38 | 1.44 | 1.44 |
| regulation of cardiac cell fate specification | 8 | 1.02 | 1.09 | 1.19 | 1.04 | 1.21 | -1.01 | 1.39 |
| negative regulation of cardiac cell fate specification | 7 | -1.04 | 1.07 | 1.14 | 1.01 | 1.15 | -1.01 | 1.35 |
| regulation of G1/S transition of mitotic cell cycle | 4 | 1.12 | 1.32 | 1.7 | 1.08 | 1.32 | 1.61 | 1.38 |
| regulation of non-canonical Wnt receptor signaling pathway | 2 | -1.01 | 1.74 | 1.54 | 1.1 | 1.38 | 1.44 | 1.44 |
| regulation of Wnt receptor signaling pathway, planar cell polarity pathway | 2 | -1.01 | 1.74 | 1.54 | 1.1 | 1.38 | 1.44 | 1.44 |
| negative regulation of DNA-dependent DNA replication | 3 | -2.66 | -1.02 | 1.24 | 1.05 | 1.35 | -2.2 | 1.46 |
| positive regulation of DNA-dependent DNA replication | 3 | 1.19 | 1.21 | 1.03 | 1.17 | 1.31 | -1.05 | 1.11 |
| regulation of cellular macromolecule biosynthetic process | 732 | 1.15 | 1.08 | 1.17 | 1.06 | 1.17 | 1.17 | 1.13 |
| negative regulation of cellular macromolecule biosynthetic process | 214 | 1.18 | 1.08 | 1.18 | 1.04 | 1.23 | 1.23 | 1.17 |
| regulation of cysteine-type endopeptidase activity | 18 | 1.12 | 1.11 | 1.23 | 1.18 | 1.35 | 1.14 | 1.01 |
| negative regulation of cysteine-type endopeptidase activity | 2 | 1.22 | -1.04 | 1.69 | -1.06 | 1.27 | -1.09 | -1.19 |
| negative regulation of G1/S transition of mitotic cell cycle | 1 | 1.02 | 1.34 | 2.07 | -1.09 | 1.41 | 2.33 | 1.77 |
| regulation of transcription initiation, DNA-dependent | 5 | 1.43 | 1.28 | 1.45 | 1.26 | 1.36 | 1.28 | 1.29 |
| positive regulation of transcription initiation, DNA-dependent | 2 | 1.1 | 1.32 | 1.52 | 1.2 | 1.24 | 1.14 | 1.45 |
| regulation of cell motility | 10 | 2.17 | -1.1 | 1.06 | -1.11 | 1.21 | 1.34 | 1.71 |
| negative regulation of cell motility | 2 | 2.1 | 1.1 | 1.42 | -1.08 | 1.39 | 1.72 | 1.26 |
| regulation of ubiquitin-specific protease activity | 1 | 1.14 | 1.09 | 1.34 | -1.05 | 1.24 | 1.35 | 2.12 |
| regulation of peptidyl-cysteine S-nitrosylation | 1 | 1.29 | 1.1 | 1.03 | 1.22 | 1.29 | 1.66 | -1.14 |
| regulation of neural precursor cell proliferation | 9 | -1.33 | 1.16 | 1.4 | 1.11 | 1.64 | 1.16 | 1.09 |
| negative regulation of neural precursor cell proliferation | 7 | -1.32 | 1.13 | 1.47 | 1.06 | 1.79 | 1.22 | 1.08 |
| positive regulation of neural precursor cell proliferation | 2 | -1.35 | 1.28 | 1.19 | 1.29 | 1.21 | -1.02 | 1.09 |
| regulation of reproductive process | 47 | 1.06 | 1.08 | 1.28 | -1.01 | 1.27 | 1.35 | 1.24 |
| negative regulation of reproductive process | 6 | 1.39 | 1.2 | 1.49 | 1.11 | 1.54 | 1.6 | 1.43 |
| positive regulation of reproductive process | 4 | -1.29 | 1.41 | 1.69 | 1.16 | 1.58 | 1.17 | 1.66 |
| negative regulation of receptor activity | 3 | 1.29 | 1.07 | 1.31 | 1.06 | 1.37 | 1.41 | 1.15 |
| positive regulation of receptor activity | 1 | -1.21 | 1.16 | 2.16 | -1.11 | 1.4 | 1.68 | 1.37 |
| regulation of epithelial cell migration, open tracheal system | 5 | 2.13 | -1.29 | -1.2 | -1.27 | 1.01 | -1.13 | 2.14 |
| regulation of DNA biosynthetic process | 1 | 1.35 | 1.23 | 1.65 | 1.22 | 1.74 | 1.39 | 1.1 |
| regulation of photoreceptor cell axon guidance | 2 | 1.11 | -1.03 | 1.73 | -1.06 | 1.28 | 1.08 | 1.11 |
| regulation of synaptic vesicle exocytosis | 3 | -1.05 | 1.27 | 1.19 | 1.07 | 1.12 | 1.29 | 1.16 |
| negative regulation of synaptic vesicle exocytosis | 1 | 1.24 | 1.25 | 1.11 | 1.13 | 1.1 | 1.34 | 1.26 |
| positive regulation of synaptic vesicle exocytosis | 1 | -1.65 | 1.56 | 1.35 | 1.0 | 1.12 | 1.4 | 1.32 |
| semaphorin-plexin signaling pathway involved in regulation of photoreceptor cell axon guidance | 2 | 1.11 | -1.03 | 1.73 | -1.06 | 1.28 | 1.08 | 1.11 |
| regulation of clathrin-mediated endocytosis | 2 | -1.04 | 1.44 | 1.65 | 1.12 | 1.42 | 2.02 | 1.59 |
| positive regulation of clathrin-mediated endocytosis | 1 | -1.65 | 1.56 | 1.35 | 1.0 | 1.12 | 1.4 | 1.32 |
| regulation of cytokinesis, actomyosin contractile ring assembly | 1 | -1.32 | 1.18 | 1.39 | 1.03 | 1.12 | 1.4 | 1.35 |
| regulation of protein neddylation | 1 | 1.11 | 1.42 | 1.38 | 1.05 | 1.25 | 1.67 | 1.2 |
| negative regulation of protein neddylation | 1 | 1.11 | 1.42 | 1.38 | 1.05 | 1.25 | 1.67 | 1.2 |
| regulation of cell proliferation involved in compound eye morphogenesis | 2 | 1.28 | 1.17 | 1.26 | 1.16 | 1.36 | 1.23 | 1.09 |
| regulation of interphase of mitotic cell cycle | 89 | 1.23 | 1.14 | 1.34 | 1.18 | 1.39 | 1.29 | 1.17 |
| regulation of histone H4-K16 acetylation | 1 | 1.8 | 1.23 | 1.54 | 1.26 | 1.37 | 1.4 | 1.76 |
| positive regulation of histone H4-K16 acetylation | 1 | 1.8 | 1.23 | 1.54 | 1.26 | 1.37 | 1.4 | 1.76 |
| regulation of transcription regulatory region DNA binding | 2 | -1.31 | 1.44 | 1.67 | 1.2 | 1.16 | 1.11 | 1.5 |
| negative regulation of transcription regulatory region DNA binding | 2 | -1.31 | 1.44 | 1.67 | 1.2 | 1.16 | 1.11 | 1.5 |
| regulation of peptidyl-lysine acetylation | 5 | 1.41 | 1.17 | 1.61 | 1.2 | 1.61 | 1.62 | 1.46 |
| negative regulation of peptidyl-lysine acetylation | 1 | 1.22 | 1.34 | 1.86 | 1.14 | 2.08 | 2.98 | 1.75 |
| positive regulation of peptidyl-lysine acetylation | 2 | 1.41 | 1.2 | 1.81 | 1.29 | 1.46 | 1.33 | 1.45 |
| regulation of steroid hormone secretion | 1 | -1.14 | 1.41 | 1.3 | 1.24 | 1.17 | 1.85 | 1.43 |
| positive regulation of cysteine-type endopeptidase activity | 14 | 1.01 | 1.14 | 1.25 | 1.19 | 1.42 | 1.19 | 1.06 |
| starch binding | 5 | -2.58 | 1.33 | -1.39 | 1.01 | -2.26 | -3.22 | 1.03 |
| regulation of RNA biosynthetic process | 671 | 1.13 | 1.08 | 1.17 | 1.06 | 1.16 | 1.15 | 1.13 |
| negative regulation of chromosome organization | 2 | 1.22 | 1.33 | 2.01 | 1.1 | 1.89 | 2.47 | 1.68 |
| positive regulation of chromosome organization | 6 | 1.14 | 1.12 | 1.43 | 1.22 | 1.52 | 1.14 | 1.15 |
| regulation of cation channel activity | 3 | 1.93 | -1.03 | 1.33 | 1.03 | 1.24 | 2.17 | 1.55 |
| positive regulation of cation channel activity | 2 | 2.64 | -1.06 | 1.57 | 1.01 | 1.45 | 3.25 | 1.94 |
| regulation of muscle cell chemotaxis toward tendon cell | 1 | 1.49 | 1.07 | 1.15 | 1.04 | -1.03 | 1.53 | -1.09 |
| negative regulation of muscle cell chemotaxis toward tendon cell | 1 | 1.49 | 1.07 | 1.15 | 1.04 | -1.03 | 1.53 | -1.09 |
| Roundabout signaling pathway involved in muscle cell chemotaxis toward tendon cell | 1 | 1.49 | 1.07 | 1.15 | 1.04 | -1.03 | 1.53 | -1.09 |
| regulation of BMP secretion | 1 | -1.14 | 1.11 | -1.09 | 1.17 | -1.12 | -1.22 | -1.05 |
| negative regulation of BMP secretion | 1 | -1.14 | 1.11 | -1.09 | 1.17 | -1.12 | -1.22 | -1.05 |
| all | 9919 | 1.03 | -1.01 | 1.01 | 1.01 | 1.05 | 1.03 | 1.01 |